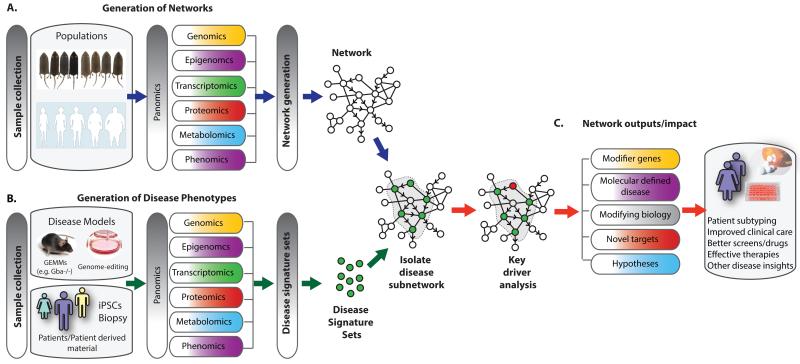
Figure 2. Studying IEM like complex disorders through adopting multi-scale omics technologies and network approaches.
We can study IEM as common disorders by taking advantage of approaches like multi-scale omics technologies and integrative network analysis and even by sharing datasets. (A) Omics data generated from samples collected in common populations of humans or model organisms can be integrated alongside public database information to generate predictive molecular networks. (B) We propose these networks can be repurposed and used as a reference or framework to associate the various IEM phenotypes, scored through multi-omics approaches on samples from IEM models and patients, to identify candidate genetic modifiers and modifying biology. For example, disease signature sets generated by various omics technologies on material derived from patients, patient-derived cells (iPSCs) or experimental model systems can be used to probe a reference network to reveal disease-associated subnetworks. (C) As these Bayesian networks have a causal predictive component they can be used to inform on key molecular drivers of the pathophysiology associated with the IEM. Genes within subnetworks can be nominated as key molecular drivers through statistical algorithms and functional and therapeutic insight can be derived through annotation of subnetwork gene members. Potential impacts of such network approaches to IEM include improving the presently poor correlation between disease severity and the primary mutated locus as well as overcoming the fundamental gap in our knowledge of disease modifying genes and biology.