Fig. 2. Eigenvector centrality scores show better correlation between co-evolution algorithms than do pairwise scores.
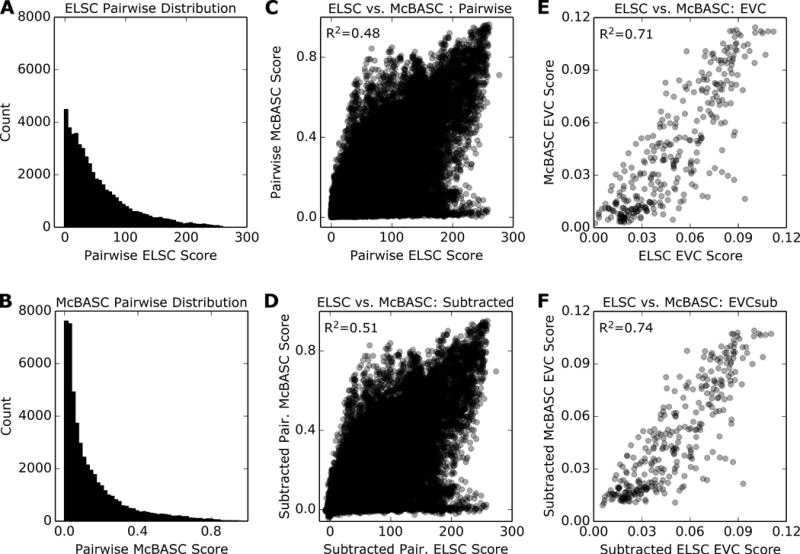
The example shown is for the aldolase family. Histograms show the distribution of co-evolutionary scores from (A) ELSC and (B) McBASC. Between these two algorithms, correlation of (C) initial (unsubtracted) or (D) subtracted pairwise co-evolution scores have modest correlations, as indicated by their Spearman R2 values. However, correlation of (E) unsubtracted or (F) subtracted EVC scores is greatly improved, as indicated by larger R2 values.
