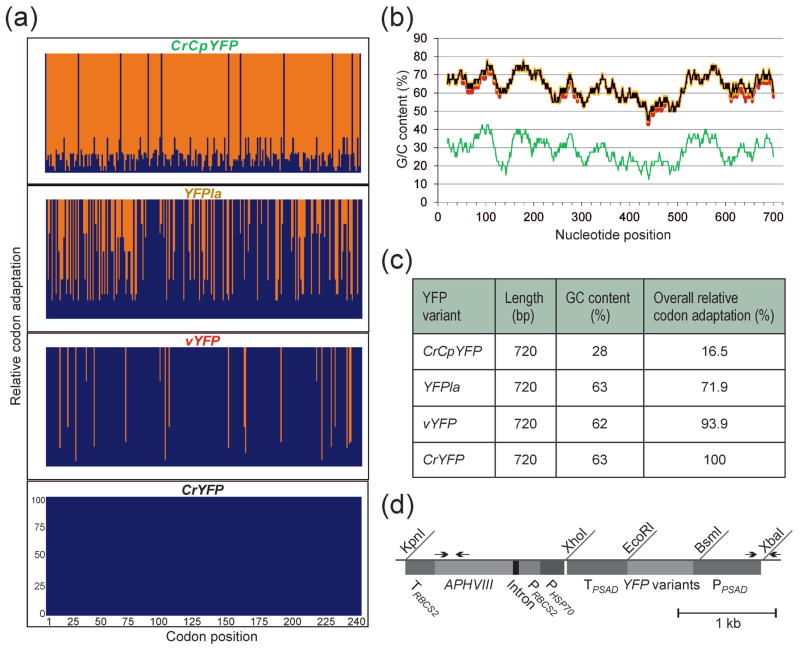
Figure 1.
YFP gene variants and their properties, and physical map of the expression vector used for nuclear transformation.
(a) Relative codon adaptation (RCA) of the different YFP gene variants compared to the nuclear genome of Chlamydomonas reinhardtii. Blue bars indicate the relative adaptation (in %) of each individual codon in the reading frames of the four gene variants. The x-axis indicates the codon numbers within the gene.
(b) GC content (in %) and its distribution over the reading frames of the four YFP gene variants. The values were determined in a sliding window of 40 bp. CrYFP is indicated in black, vYFP in red, YFPla in orange and the A/T-rich variant CrCpYFP in green (cf. colors in panel a).
(c) Overall properties of the different YFP gene variants. All variants have the same size and identical amino acid sequence, but they are highly variable in their GC content and codon usage (RCA).
(d) Physical map of the transformation vector used for expression of the YFP gene variants in Chlamydomonas. The YFP genes were cloned into a PSAD expression cassette (Fischer and Rochaix, 2001) using the restriction sites BsmI and EcoRI. The paromomycin resistance gene APHVIII serves as selectable marker and is driven by a hybrid promoter consisting of fused expression elements from the HSP70A gene (PHSP70) and the RBCS2 gene (PRBCS2). Arrows indicate primers used for PCR analysis of transformed strains. PPSAD: promoter from the PSAD gene; TPSAD: terminator from the PSAD gene.