Abstract
Epithelioid Sarcoma (ES) is a rare neoplasm uniquely comprised of cells exhibiting both mesenchymal and epithelial features. Having propensity for local and distant recurrence, it poses a diagnostic dilemma secondary to pathologic complexity. Patients have dismal prognosis due to lack of effective therapy. HDAC inhibitors (HDACi) exhibit marked anti-tumor effects in various malignancies. The studies here demonstrate that pan-HDAC inhibitors constitute novel therapeutics versus ES.
Human ES cells (VAESBJ, HS-ES, Epi-544) were studied in preclinical models to evaluate HDACi effects. Immunoblot and RT-PCR were used to evaluate expression of acetylated tubulin, histones H3/H4, EZH2 upon HDACi. MTS and clonogenic assays were used to assess the impact of HDACi on cell growth. Cell culture assays were used to evaluate the impact of HDACi and EZH2-specific siRNA inhibition on cell cycle progression and survival. Unbiased gene array analysis was used to identify the impact of HDACi on ES gene expression. Xenografts were used to evaluate ES tumor growth in response to HDACi.
HDAC inhibition increased target protein acetylation and abrogated cell growth and colony formation in ES cells. HDACi induced G2-cellcycle arrest and marked apoptosis, and reduced tumor growth in xenograft models. HDACi induced widespread gene expression changes, and EZH2 was significantly down-regulated. EZH2 knockdown resulted in abrogated cell growth in vitro.
Implications
The current study suggests a clinical role for HDACi in human ES, Which when combined with EZH2 inhibitors could serve as a novel therapeutic strategy for ES patients. Future investigations targeting specific HDAC isoforms along with EZH2 may potentially maximizing treatment efficacy.
Keywords: Epithelioid sarcoma (ES), HDAC inhibitor (HDACi), Enhancer of zeste homolog 2 (EZH2), Abexinostat (Abx), The Molecular Signatures Database (MSigDB)
Introduction
Epithelioid sarcoma (ES) is a rare soft tissue malignancy of unknown origin, exhibiting unique histologic characteristics reminiscent of both mesenchymal and epithelial differentiation (1, 2). This rare subtype of soft tissue sarcoma (STS) comprises <1% of cases and exhibits marked propensity for lymphatic spread (3). Typically affecting young adults, it is classified into two distinct histopathological subtypes: conventional classic distal type, comprised of epithelioid tumor cells in nodular morphology surrounding an area of central necrosis, and the more recently identified aggressive proximal or axial type, typically exhibiting an epithelioid or rhabdoid morphology (1, 4, 5). To date, complete surgical resection of localized tumors remains the primary therapy; locally advanced and metastatic ES are usually unresectable, chemoresistant, and fatal (6). Emerging evidence suggests that epigenetic alterations in the integrity of the nucleosomal structure can lead to various oncogenic events, including tumor progression. Studies have shown correlation between several molecular targets and pathways linked to the aggressiveness and metastatic potential of ES (notably EGFR/mTOR deregulation; [7]), and the loss of INI-1 observed in approximately 95% of ES (2). The SMARCB1/INI1 gene encodes for a subunit of SWItch/Sucrose Non-Fermentable (SWI/SNF) chromatin remodeling complex; it is reported to function as a tumor suppressor gene in infantile malignant rhabdoid tumors (8,9).
Histone deacetylases (HDAC) are a family of enzymes that deacetylate lysines on core histone and non-histone proteins (10). HDACs play a significant epigenetic role by inducing changes in chromatin structure or functioning as a “cog” in a protein complex.
Eleven mammalian HDAC isoforms have been identified and categorized into four classes based on sequence identity and domain organization (10). Many broad-spectrum compounds have been developed that target numerous HDAC isoforms, and are currently being evaluated for treatment of various malignancies (11). HDAC inhibitors (HDACi) display an array of anti-cancer effects including tumor cell growth inhibition, cell death, anti-angiogenesis in vitro and in vivo (12). Previously, our group demonstrated remarkable anti-STS effects of HDAC inhibition alone and in combination with chemotherapy (13); effects of HDAC inhibition alone in NF1-associated malignant peripheral nerve sheath tumors (MPNST) as well as combined HDACi and autophagy inhibition in vitro and in vivo (14).
In this study, we explored HDAC inhibitory drugs in ES, demonstrating that their mechanisms of action lead to abrogated cell growth, proliferation, and increased apoptosis in both in vitro and in vivo in preclinical contexts.
Materials and Methods
Cell lines and reagents
Human ES cell lines used for this study: VAESBJ, HS-ES, Epi-544. VAESBJ was obtained from the American Type Culture Collection; HS-ES was obtained from Hiroshi Sonobe (Kochi Medical School, Nankoku, Japan); Epi-544 was established in our laboratory (2) (All three are p53 wild-type as sequenced in our lab). All ES cell lines were subjected to short tandem repeat (STR) analysis. STR DNA fingerprinting was done by the Cancer Center Support Grant-funded Characterized Cell Line core UTMDACC (MD Anderson Cancer Center), NCI #CA016672. Cell lines received from collaborators and ATCC were tested upon arrival to our lab. Stock of STR confirmed cell lines were frozen at large quantities to have an original stock of cells. Cell lines were passed no more than 4-50 passages, after which, researchers utilized cells from original stocks. ES cells were cultured in DMEM 1× supplemented with 10% FBS (Life Technologies). Pan-HDAC inhibitor abexinostat was obtained from Pharmacyclics; suberoylanilide hydroxamic acid (SAHA) and MS-275 were acquired from Dr. David McConkey, MD Anderson (initially obtained from Syndax Pharmaceuticals, Inc.) and dissolved in DMSO to create stock solutions. All additional dilutions were completed using the respective cell culture medium for each cell line. Commercial antibodies were used for Western blot analysis or immunohistochemical detection: acetylated H3, acetylated H4 (Millipore); acetylated tubulin (Sigma); EZH2, caspase-3 (Cell Signaling); β-actin (Santa Cruz); PARP, Survivin, HDAC1, HDAC2, HDAC3 (Abcam); Ki67 (MIB-1). Secondary antibodies included horseradish peroxidase–conjugated (WB: anti-rabbit, anti-mouse [Santa Cruz]; IHC: Universal kit HRP; Biocare Medical). Hoechst 33342 (Polysciences) was used for immunofluorescence experiments.
Growth related studies: MTS, clonogenicity, cell cycle, apoptosis
MTS, clonogenicity, cell cycle, and annexin V assays were performed as previously described (13). Apoptosis was measured using the Apoptosis Detection kit I (BD Biosciences) as per manufacturer's recommendations. Further information is available as Supplementary Data.
WB analyses
Western blot analyses were performed by standard methods (13).
qRTPCR
Reverse transcription-PCR/quantitative reverse transcription-PCR. These assays were conducted as previously described (13). EZH2 and actin PCR primers for quantitative reverse transcription-PCR (qRTPCR) were purchased from Sigma.
Gene arrays
Gene expression profiling was conducted using an Illumina gene array. Target preparation of the samples were conducted using Illumina TotalPrep RNA kit (Ambion, Cat.# AMIL1791). 200 ng total RNA was used in each amplification reaction. The TotalPrep™ RNA Amplification Kit is based on the RNA amplification protocol developed in the laboratory of James Eberwine (26). The procedure consists of reverse transcription with an oligo (dT) primer bearing a T7 promoter using Ambion's proprietary ArrayScript™, a reverse transcriptase (RT) engineered to produce higher yields of first strand cDNA than wild type enzymes. ArrayScript™ catalyzes the synthesis of virtually full-length cDNA, which is the best way to ensure production of reproducible microarray samples. The cDNA then undergoes second strand synthesis and clean-up to become a template for in vitro transcription with T7 RNA Polymerase. To maximize cRNA yield, Ambion's proprietary MEGAscript® in vitro transcription (IVT) technology, along with biotin UTP (provided in the kit), is used to generate hundreds to thousands of biotinylated, antisense RNA copies of each mRNA in a sample. Hybridization was carried out following Illumina's direct hyb gene expression protocol. We hybridized 750 ng of amplified biotinylated cRNA onto Illumina's HumanHT12v4 gene expression arrays. The arrays were washed according to manufacturer's specifications and imaged using Illumina iScan Scanner. Raw feature intensity files (.idat) were generated using Illumina's GenomeStudio software. Quantile normalized array values were log-transformed. Array data were deposited in the Gene Expression Omnibus (accession GSE66800). SigTerms (27) software was used to search the The Molecular Signatures Database (MSigDB, version 3.0) for significantly enriched pathway associations.
siRNA Knockdown
siRNAs (20 nM pools targeting EZH2, HDAC1, HDAC2, HDAC3, and control non-targeting constructs; Thermo Scientific, Waltham, MA) were introduced into cells using X-treme Gene per manufacturer's instructions (Roche, Mannheim, Germany). Briefly, 2×105 cells were plated in each well of a six-well plate and incubated overnight. A mixture of siRNA (20 nM) and X-treme Gene (6 μl) diluted in 100 μl Dulbecco's modified Eagle medium (DMEM) was added for 24 hr, followed by incubation in regular medium. Cells were harvested at indicated time points for specific experiments.
In vivo therapeutic experiments
All animal procedures and care were approved by the MD Anderson Cancer Center Institutional Animal Care and Usage Committee. Animals received humane care as per the Animal Welfare Act and the NIH “Guide for the Care and Use of Laboratory Animals”. For experiments evaluating effect of abexinostat monotherapy on local tumor growth trypan blue staining confirmed viable VAESBJ cells. Cell suspensions (2×106) were injected subcutaneously into the flank of 6-7 week old female hairless SCID mice and growth was measured twice weekly. After establishment of palpable lesions (average diameter ∼4-5 mm), mice were randomly assigned to receive either vehicle control or abexinostat (25 mg/kg BID). Abexinostat was solubilized in 50mM sodium lactate, pH4.2 and administered i.p. twice daily, five days/week. Treatment continued until mice in control group mandated euthanasia. Tumors were resected, weighed, and fixed in formalin and paraffin-embedded for immunohistochemical studies. Immunohistochemistry on paraffinized, xenograft-derived specimen was conducted as previously described (7).
Statistical analyses
Cell culture-based assays were repeated at least 3×; mean and SD was calculated and shown in the figures. Cell lines were examined separately. For outcomes that were measured at a single time point, two-sample t-tests were used to assess the differences for independent data. Differences in xenograft growth in vivo were analyzed with linear mixed effects models to take account of the correlations among observations from the same animal as the tumor volume was measured over time. Tumor weight between groups was compared with Analysis of Variance (ANOVA) P-value < 0.05 was considered as significant.
Results
HDAC inhibitors abrogate ES cell growth in vitro
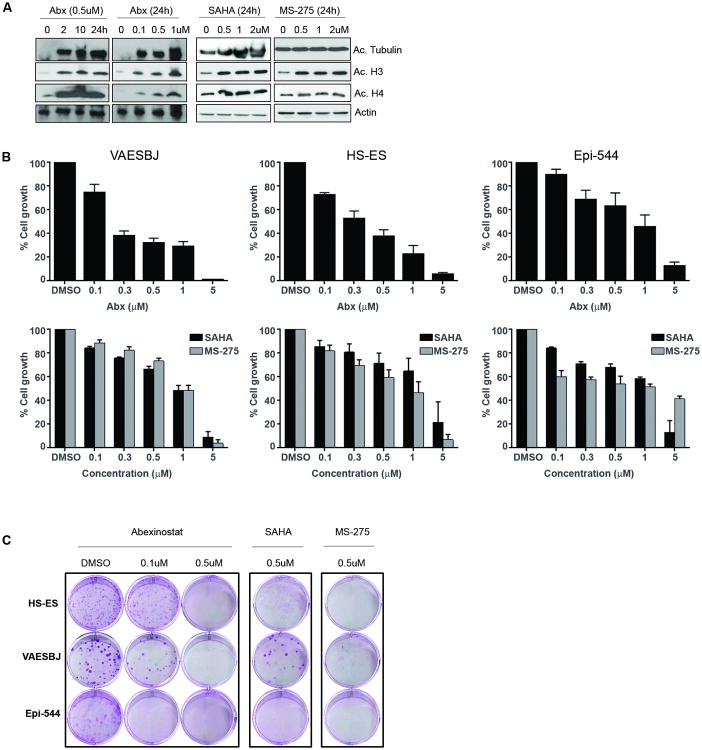
Pan-HDACi abexinostat (Abx) was utilized in all experiments; pan-HDACis SAHA and MS-275 were used as comparative compounds to show that abexinostat effects were not unique to a single HDACi agent. Previously, we demonstrated anti-tumor efficacy of abexinostat in numerous STS histologies, and dose and administration regiment for this study were based on previously published data (13, 14). As anticipated, all three compounds induced acetylation of target proteins in all ES cell lines tested (Fig. 1A). The effects of HDAC inhibition on ES cell growth and colony formation were evaluated. Abexinostat (96 hrs) induced a time- and dose-dependent decrease in VAESBJ, HS-ES, and Epi-544 cell growth (Fig. 1B). All ES cell lines tested also exhibited growth inhibition in response to SAHA and MS-275 treatment. Similarly, HDAC inhibition decreased ES cell colony formation compared with DMSO-treated controls (Fig. 1C and Supplementary Fig. S1).
Figure 1. HDACis inhibit the growth of ES cells in vitro.
(A) Pan-HDAC inhibition induces target protein acetylation in VAESBJ cells. VAESBJ cells were tested with HDAC inhibitors (abexinostat, SAHA, MS-275) to confirm protein target acetylation of tubulin, histone 3, and histone 4. All three compounds increased target acetylation in a dose- and time-dependent manner; as expected, MS-275 did not increase tubulin acetylation due to its class I HDAC specificity. (B) HDACi effect on growth was determined using MTS assay. HDACi abrogated ES cell line growth, where abexinostat exhibited a greater effect over SAHA and MS-275. This effect may be due to the higher affinity of HDAC isoforms of abexinostat compared to SAHA and MS-275. (C) HDACi significantly reduced colony formation of ES cells (10 days). Graphs of triplicate clonogenic assays are depicted in supplemental data.
HDACi induces G2 cell cycle arrest and apoptosis in ES cells
We evaluated whether HDACi-induced ES growth inhibition could be due to effects on ES cell cycle progression and/or induction of apoptosis. ES cell lines were treated with increasing doses (0.5, 1 μM) of abexinostat for 48 hours; samples were then analyzed using propidium iodide staining/FACS analysis. Significant dose-dependent increases (P < 0.05) in G2 cell cycle arrest was observed in abexinostat–treated cells compared with DMSO–treated controls (Fig. 2A). Cell cycle analysis also revealed an increase in the sub-G1 fraction in abexinostat-treated cells, suggesting HDACi-induced apoptosis. To identify potential HDACi-induced apoptosis, the effects of abexinostat on phosphoatidylserine exposure was tested by Annexin V staining and FACS analysis. A significant increase in HDACi-induced apoptosis was observed in all three cell lines (P < 0.05; Fig. 2B). Abexinostat-induced increases in apoptosis markers, cleaved PARP, and cleaved caspase 3 were observed in Western blot analysis (Fig. 2C). Together, these data demonstrate the in vitro monotherapeutic efficacy of pan-HDACi in ES cells.
Figure 2. HDACi induces G2 cell cycle arrest and apoptosis in ES cells in vitro.
(A) Propidium iodide-FACS analysis abexinostat (0.5, 1 μM/48 h) induced a significant increase in the G2/M phase (48 h) in tested ES cells (P < 0.05). VAESBJ exhibited the strongest G2/M phase shift after abexinostat treatment compared to HS-ES and Epi-544. (B) Annexin V/PI-FACS analysis demonstrated a significant increase in abexinostat-induced (1 μM/48 h) apoptosis was observed in all three ES cell lines. (C) Western blot analysis confirmed Annexin V data where abexinostat (1 μM/48 h) induced an increase in apoptosis markers cleaved PARP and cleaved caspase 3.
HDACi abrogates ES xenograft growth
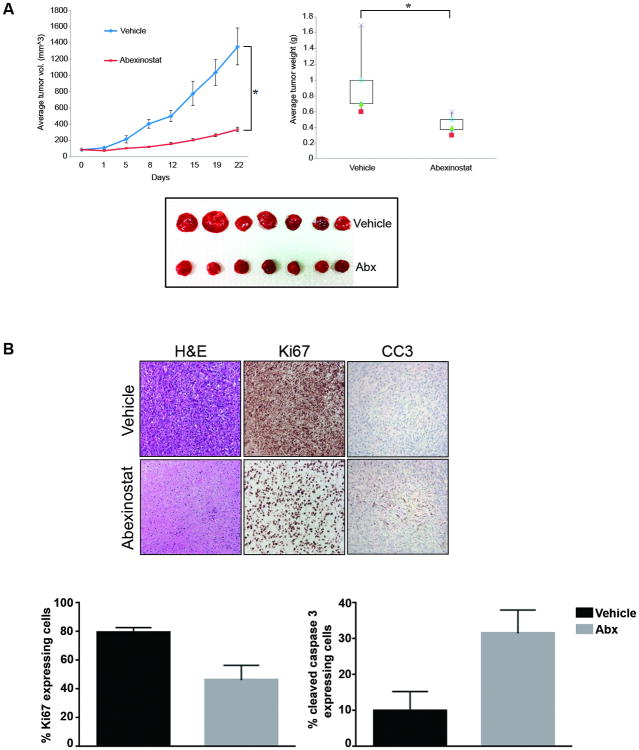
We next assessed the in vivo effect of HDACi on ES using human VAESBJ xenograft tumors. Mice were injected with viable VAESBJ cells s.c. (2×106); therapy was administered once tumors grew to a mean diameter of 0.5 cm. No apparent side effects were noticed in mice receiving vehicle or abexinostat during the course of therapy. Results (Fig. 3A) demonstrate abrogation of tumor growth in mice treated with low dose abexinostat (25 mg/kg/BID) with significantly reduced tumor volume over time compared to vehicle control (P < 0.0001). Moreover, abexinostat induced a significant decrease in tumor weight compared to vehicle-treated mice as well (P < 0.0001). Immunohistochemical analysis of the tumor samples exhibit a significant decrease in proliferation (Ki67, P < 0.05) and increase in apoptosis (cleaved caspase 3, P < 0.05; Fig. 3B). Together, in vitro and in vivo data suggests that HDAC inhibition is potentially effective in ES.
Figure 3. HDACi abrogates ES xenograft growth.
(A) SCID mice with VAESBJ xenografts were treated with vehicle or abexinostat (25 mg/kg/BID). Tumor volumes, weight, and images are depicted demonstrating significant abexinostat-induced tumor growth inhibition.
(B) Immunohistochemical analysis of vehicle and abexinostat-treated tumor sections demonstrate reduction in cell proliferation (Ki67; P < 0.05) and increased apoptosis (CC3; P < 0.05).
HDACi induces gene expression signature in ES
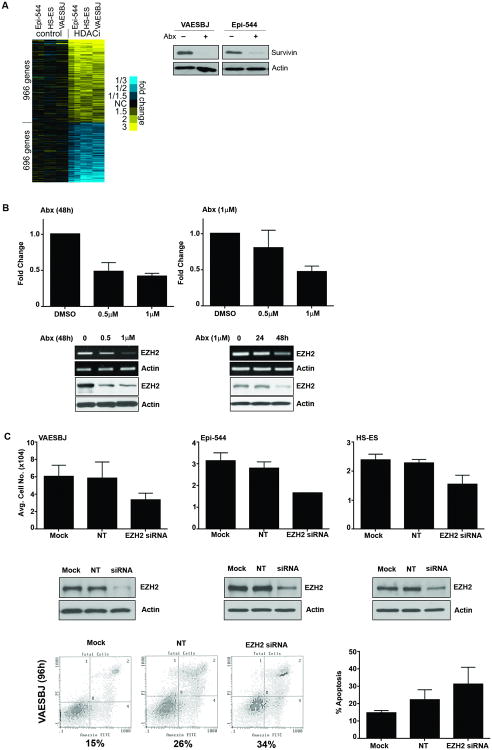
To identify possible HDACi-induced gene changes in ES cells, ES cells were treated with abexinostat (0.5 μM, 24 h) and gene expression profiles of each cell line (DMSO- and abexinostat-treated samples) were conducted using an Illumina gene array. Array analysis identified an increase in 966 genes in abexinostat-treated samples vs DMSO control and a decrease in 696 genes in abexinostat-treated samples vs DMSO control (Fig. 4A, P < 0.01, t-test, fold changes > 1.5). Among the differentially expressed genes, BIRC5/Survivin protein expression was assessed to confirm the array (Fig. 4A); this protein being of particular interest due to its well-established role in apoptosis (15).
Figure 4. HDACi induces widespread changes in gene expression and reduces EZH2 expression in ES.
(A) By gene array analysis, top differentially expressed genes (P < 0.01, t-test, fold change > 1.5) in ES cells treated with abexinostat versus control (DMSO)-treated cells. Gene array results confirmed include decreased Survivin protein expression in response to abexinostat. (B) In ES, PCI2 represses EZH2 expression in a dose- and time-dependent manner. (C) EZH2 siRNA knockdown reduces ES cell line growth.
In order to carry out pathway analysis to identify global association, our gene array results were run against The Molecular Signatures Database (MSigDB), from which a significant number of genes were found to be commonly up- and down-regulated in our gene sets and in the NUYTTEN_EZH2_TARGETS gene sets (Supplementary Table 1 S1, [16]). In following up these bioinformatics results, abexinostat reduced EZH2 expression in a time- and dose-dependent manner in ES cells (Fig. 4B and Supplementary Fig. S2).
To identify if EZH2 plays a role in ES cell growth, mock (transfection reagent alone), non-target control siRNA (NT) and EZH2 siRNA transfection was used. Confirmation of EZH2 by siRNA knockdown was demonstrated by western blot which show decrease in EZH2 protein expression. EZH2 knockdown induced ES cell growth inhibition: counted cell numbers were significantly lower in siRNA treated cells vs mock or non-target siRNA controls; knockdown of EZH2 in VAESBJ also induced an increase in apoptosis (Fig. 4C). These data demonstrate a potential role for EZH2 in ES, and suggests that HDACi-induced EZH2 regulation may possibly be relevant in HDACi-induced anti-ES effects.
Conclusions
Our current study highlights a unique ES-associated epigenetic crosstalk between HDACs and EZH2 that is of potential translational and clinical importance. HDAC inhibitors have gained greater focus in anti-cancer therapy because their deacetylation inhibition of both histones and non-histone cellular proteins facilitates important transcriptional changes in target genes, especially those involved in differentiation, cellular proliferation and apoptosis (12). Various HDACis cause numerous effects in different malignancies, including induction of differentiation, cell death, cell-cycle arrest, altered cell migration, and apoptosis (11-14). Our findings suggest an important oncogenic suppression: upon treatment with pan-HDAC inhibitors, abrogation of colony formation and reduced cell growth was apparent in three ES cell lines tested (VAESBJ, HS-ES, Epi-544). We evaluated the effects of pan-HDACis on ES growth in relation to cell cycle progression and induction of apoptosis, demonstrating significant dose-dependent increases in G2/M cell cycle arrest and increased apoptosis. Similar anti-ES effects in response to HDAC inhibition were also seen in vivo.
EZH2 is a histone methyltransferase that catalyzes methylation of histone H3 lysine 27; along with SUZ12, EED, and RbAp46/48 it forms the Polycomb Repressive Complex 2 (PRC2; [17]). EZH2 is overexpressed in many tumors, including high-grade and advanced-stage breast, prostate, and lung cancers (18). It mainly serves to maintain the transcriptional repressive state of genes over successive cell generations (17). In our study, we saw a significant reduction in cell growth and proliferation when we targeted EZH2 via siRNA. EZH2 knockdown also showed an increase in apoptosis in VAESBJ. Previous studies have shown Polycomb-group proteins (PcG), particularly PRC2, mediating repression of gene activity by their involvement with HDAC proteins (19, 20). Moreover, we found that some HDAC isoforms have a more profound role than others regarding the regulation of EZH2 (Supplementary Fig. S3); specifically, we observed that HDAC 1/2 have greater effects on the protein level expression of EZH2. When we knocked down HDAC1, we saw an induced reduction in EZH2, as well as similar results in HDAC2 knockdown experiments. When we induced knockdown of HDAC3; however, we did not see changes in levels of EZH2 protein. Although a more inclusive study examining molecular interactions between HDACs and EZH2 is needed, it is likely an important function of the HDAC activity. Whether the mechanism by which HDAC inhibitors deplete EZH2 is at the transcriptional or post-transcriptional level, or perhaps via increased protein degradation remains to be elucidated.
It will be clinically relevant to explore rational targeted therapeutic combinations to achieve more potent antitumor effects while overcoming initial ES therapeutic resistance (7). Identifying additional ES-associated targetable molecular deregulations in combination with HDAC inhibitors would be of great therapeutic utility. Tumors frequently exhibit alterations in the expression and composition of proteins that regulate cellular proliferation and death. Yamaguchi et al. demonstrated significant synergistic inhibition of gallbladder carcinoma proliferation via HDAC inhibition and repression of EZH2 (21). Fiskus and colleagues observed similar downregulation of EZH2 upon HDAC inhibition in human acute leukemia cells (22).
Taken together, EZH2 and its cooperation with HDACs may be related to the tumorigenesis of epithelioid sarcoma, as has been reported in other types of cancers (23-25). Although the limitation of having only a small cohort of ES cell lines available for testing suggests caution, these findings support further investigation of targeted therapeutics versus HDAC isoforms with combined EZH2 inhibitors, a novel strategy which could ultimately contribute to meaningful clinical options for patients burdened by ES.
Supplementary Material
Acknowledgments
The authors thank David J. McConkey for provided SAHA and MS-275 to supplement this study. We thank Yiqun Zhang for technical assistance with gene array analysis. We thank Pharmacyclics for providing abexinostat.
Footnotes
Competing interests: The authors declare that they have no competing interests.
References
- 1.Gasparini P, Facchinetti F, Boeri M, Lorenzetto E, Livio A, Gronchi A, et al. Prognostic determinants in epithelioid sarcoma. Eur J Cancer. 2011;47:287–95. doi: 10.1016/j.ejca.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 2.Sakharpe A, Lahat G, Gulamhusein T, Liu P, Bolshakov S, Nguyen T, et al. Epithelioid Sarcoma and Unclassified Sarcoma with Epithelioid Features: Clinicopathological Variables, Molecular Markers, and a New Experimental Model. Oncologist. 2011;16:512–22. doi: 10.1634/theoncologist.2010-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Visscher SA, van Ginkel RJ, Wobbes T, Veth RP, Ten Heuvel SE, Suurmeijer AJ, et al. Epithelioid sarcoma: Still an only surgically curable disease. Cancer. 2006;107:606–612. doi: 10.1002/cncr.22037. [DOI] [PubMed] [Google Scholar]
- 4.Guillou L, Wadden C, Coindre JM, Krausz T, Fletcher CD. “Proximal-type” epithelioid sarcoma, a distinctive aggressive neoplasm showing rhabdoid features. Clinicopathologic, immunohistochemical, and ultrastructural study of a series. Am J Surg Pathol. 1997;21:130–146. doi: 10.1097/00000478-199702000-00002. [DOI] [PubMed] [Google Scholar]
- 5.Hasegawa T, Matsuno Y, Shimoda T, Umeda T, Yokoyama R, Hirohashi S. Proximal-type epithelioid sarcoma: A clinicopathologic study of 20 cases. Mod Pathol. 2001;14:655–663. doi: 10.1038/modpathol.3880368. [DOI] [PubMed] [Google Scholar]
- 6.Wolf PS, Flum DR, Tanas MR, Rubin BP, Mann GN. Epithelioid sarcoma: the University of Washington experience. Am J Surg. 2008;196:407–12. doi: 10.1016/j.amjsurg.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 7.Xie X, Ghadimi MP, Young ED, Belousov R, Zhu QS, Liu J, et al. Combining EGFR and mTOR Blockade for the Treatment of Epithelioid Sarcoma. Clin Cancer Res. 2011;17:5901–12. doi: 10.1158/1078-0432.CCR-11-0660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–6. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 9.Biegel JA, Zhou JY, Rorke LB, Stenstrom C, Wainwright LM, Fogelgren B. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 1999;59:74–9. [PubMed] [Google Scholar]
- 10.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737–49. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dokmanovic M, Clarke C, Marks PA. Histone deacetylases inhibitors: overview and perspectives. Mol Cancer Res. 2007;5:981–9. doi: 10.1158/1541-7786.MCR-07-0324. [DOI] [PubMed] [Google Scholar]
- 12.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–84. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 13.Lopez G, Liu J, Ren W, Wei W, Wang S, Lahat G, et al. Combining PCI-24781, a novel histone deacetylase inhibitor, with chemotherapy for the treatment of soft tissue sarcoma. Clin Cancer Res. 2009;15:3472–83. doi: 10.1158/1078-0432.CCR-08-2714. [DOI] [PubMed] [Google Scholar]
- 14.Lopez G, Torres K, Liu J, Hernandez B, Young E, Belousov R, et al. Autophagic survival in resistance to histone deacetylase inhibitors: novel strategies to treat malignant peripheral nerve sheath tumors. Cancer Res. 2011;71:185–96. doi: 10.1158/0008-5472.CAN-10-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mita AC, Mita MM, Nawrocki ST, Giles FJ. Survivin: Key Regulator of Mitosis and Apoptosis and Novel Target for Cancer Therapeutics. Clin Cancer Res. 2008;14:5000–5. doi: 10.1158/1078-0432.CCR-08-0746. [DOI] [PubMed] [Google Scholar]
- 16.Nuytten M, Beke L, Van Eynde A, Ceulemans H, Beullens M, Van Hummelen P, et al. The transcriptional repressor NIPP1 is an essential player in EZH2-mediated gene silencing. Oncogene. 2008;27:1449–60. doi: 10.1038/sj.onc.1210774. [DOI] [PubMed] [Google Scholar]
- 17.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–35. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat Genet. 1999;23:474–8. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- 20.Richter GH, Plehm S, Fasan A, Rossler S, Unland R, Bennani-Baiti IM, et al. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. PNAS. 2009;160:5324–9. doi: 10.1073/pnas.0810759106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamaguchi J, Sasaki M, Sato Y, Itatsu K, Harada K, Zen Y, et al. Histone deacetylase inhibitor (SAHA) and repression of EZH2 synergistically inhibit proliferation of gallbladder carcinoma. Cancer Sci. 2009;101:355–62. doi: 10.1111/j.1349-7006.2009.01387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fiskus W, Pranpat M, Balasis M, Herger B, Rao R, Chinnaiyan A, et al. Histone deacetylase inhibitors deplete enhancer of zeste 2 and associated polycomb repressive complex 2 proteins in human acute leukemia cells. Mol Cancer Ther. 2006;5:3096–104. doi: 10.1158/1535-7163.MCT-06-0418. [DOI] [PubMed] [Google Scholar]
- 23.Tong ZT, Cai MY, Wang XG, Kong LL, Mai SJ, Liu YH, et al. EZH2 supports nasopharyngeal carcinoma cell aggressiveness by forming a co-repressor complex with HDAC1/HDAC2 and Snail to inhibit E-cadherin. Oncogene. 2012;31:583–94. doi: 10.1038/onc.2011.254. [DOI] [PubMed] [Google Scholar]
- 24.Chen H, Tu SW, Hsieh JT. Down-regulation of human DAB2IP gene expression mediated by polycomb Ezh2 complex and histone deacetylase in prostate cancer. J Biol Chem. 2005;280:22437–44. doi: 10.1074/jbc.M501379200. [DOI] [PubMed] [Google Scholar]
- 25.Tonini T, Bagella L, D'Andrilli G, Claudio PP, Giordano A. Ezh2 reduces the ability of HDAC1-dependent pRb2/p130 transcriptional repression of cyclin A. Oncogene. 2004;23:4930–7. doi: 10.1038/sj.onc.1207608. [DOI] [PubMed] [Google Scholar]
- 26.Van Gelder RN, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. PNAS. 1990;87:1663–7. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Creighton CJ, Nagaraja AK, Hanash SM, Matzuk MM, Gunaratne PH. A bioinformatics tool for linking gene expression profiling results with public databases of microRNA target predictions. RNA. 2008;14:2290–6. doi: 10.1261/rna.1188208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.