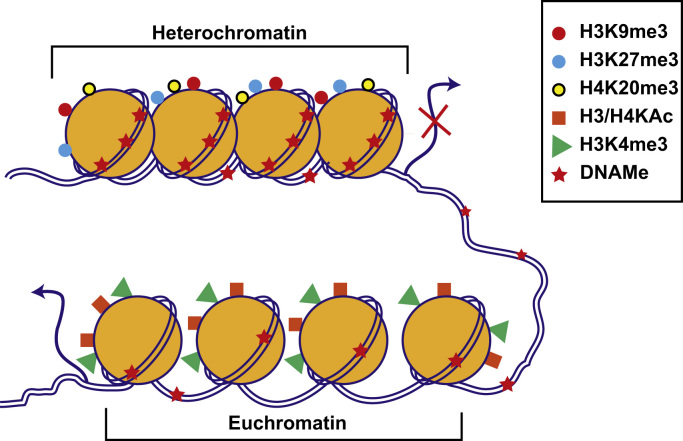
Figure 1.
Chromatin structure and function. Chromatin is made up of repeating units of nucleosomes consisting of 146 bp DNA wrapped around dimers of four histone proteins (H2A, H2B, H3, and H4). The exposed amino-terminal tails of nucleosomal histones are subjected to post-translational modifications. Combinatorial effects of histone modifications and DNAMe regulate the chromatin structure between transcriptionally silent “heterochromatin” and active “euchromatin.” Enrichment of promoter DNAMe and histone modifications such as H3K9me3, H3K27me3, and H4K20me3 promote nucleosome condensation to repress transcription (heterochromatin). On the other hand, histone modifications H3 or H4 KAc (H3/H4KAc) and H3K4me promote open chromatin formation and increase accessibility to the transcription machinery, leading to active transcription (euchromatin). Other histone modifications such as Ser/Thr phosphorylation, ubiquitination and SUMOylation, and non-coding RNAs including microRNAs also regulate chromatin structure and function (not shown). Genome-wide patterns of DNAMe and histone modifications are referred to as the “epigenome.” Its response to internal and external signals regulates gene expression involved in diverse biological processes and disease conditions. DNAMe, DNA methylomes; KAc, lysine acetylation.