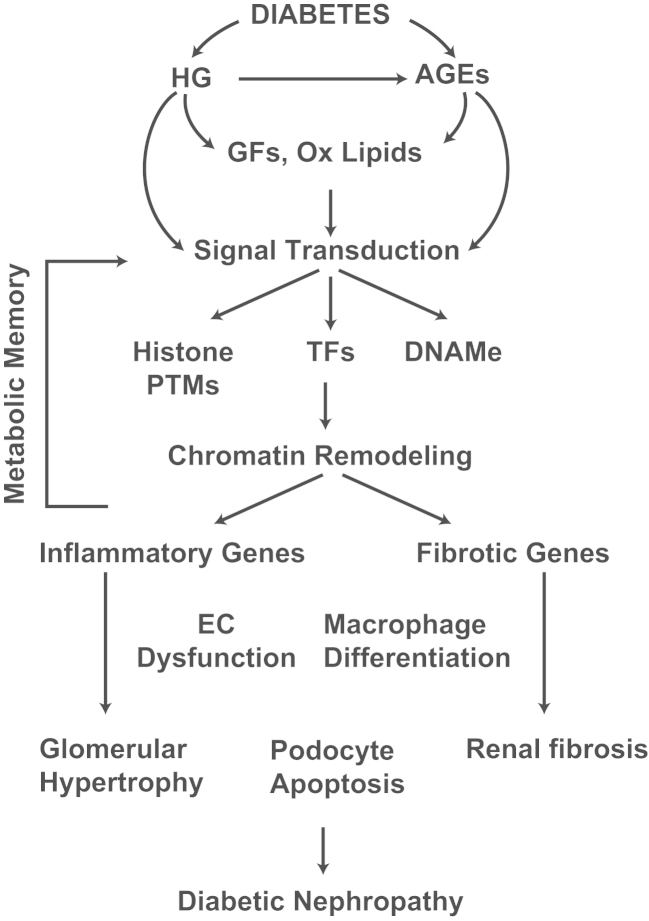
Figure 2.
Diabetic nephropathy and metabolic memory. Signal transduction mediated by factors associated with the diabetic milieu (HG, AGEs, growth factors, and oxidized lipids) induces alterations in epigenetic modifications (histone PTMs and DNAMe) and changes in chromatin remodeling. This leads to pathologic gene expression (inflammatory genes and fibrotic genes) in collaboration with key TFs in monocytes/macrophages, vascular and renal cells. Persistence of these epigenetic modifications even after normalizing glucose levels can play an important role in “metabolic memory” and gene expression implicated in long-lasting diabetic complications including nephropathy. AGEs, advanced glycation end products; DNAMe, DNA methylomes; EC, endothelial cell; GF, growth factor; HG, high glucose; Ox lipids, oxidized lipids; PTM, post-translational modification; TF, transcription factor.