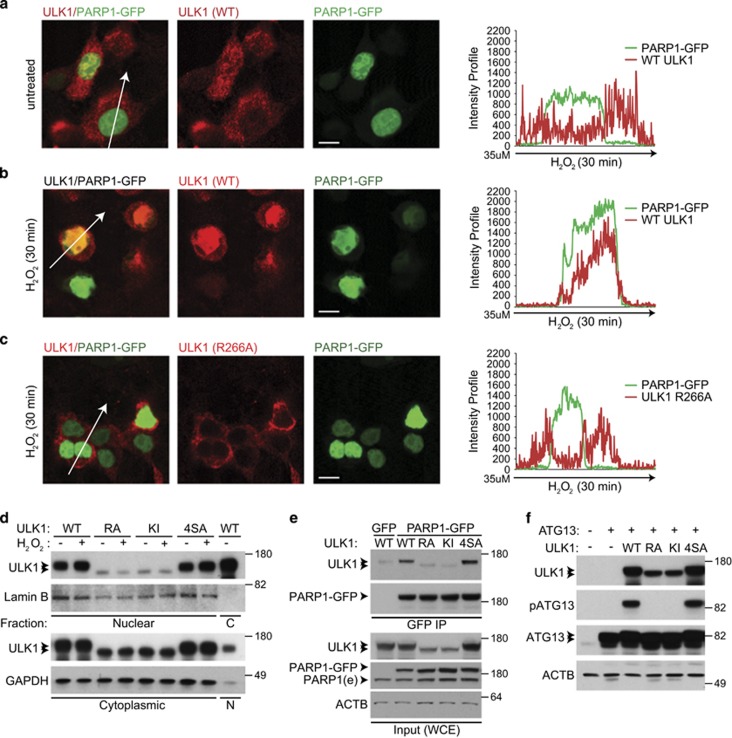
Figure 6.
ULK1 kinase activity is required for nuclear localization of ULK1 and its interaction with PARP1. (a and b) The 293T cells cotransfected with PARP1-GFP and untagged WT ULK1 were incubated in complete media (CM) in the absence (a) or the presence (b) of H2O2 for 30 min. The percentage of cells showing clear nuclear localization of ULK1 increased from 1% to nearly 30% after H2O2 treatment. Representative images and line scans indicate the degree of colocalization between PARP1-GFP and ULK1 in cells treated with or without H2O2. (c) The 293T cells cotransfected with PARP1-GFP and the nuclear localization-defective R266A-ULK1 mutant were treated for 30 min with 500 μM H2O2 before imaging. Representative images and a line scan show the degree of colocalization of R266A-ULK1 and PARP1-GFP, highlighting the nuclear localization defect of the R266A mutant. Scale bars=10 μM. (d) Representative immunoblots of nuclear and cytoplasmic fractions prepared from 293T cells transfected with WT or mutant ULK1 (R266A, KI, or 4SA) and incubated in CM with or without 500 μM H2O2 for 30 min. The levels of WT ULK1 and the 4SA mutant in nuclear fractions increase after H2O2 treatment, but those of the KI and R66A mutants do not. (e) Whole-cell extracts prepared from 293 T cells cotransfected with PARP1-GFP and untagged WT or mutant (KI, R266A, or 4SA) ULK1 were subjected to IP using anti-GFP-conjugated beads before SDS-PAGE and immunoblot analyses. Immunoblot analyses demonstrate a decrease in PARylated proteins in cells transfected with the R266A mutant. (f) The 293T cells were transfected with HA-tagged ATG13 and the indicated ULK1 expression constructs. Whole-cell extracts were analyzed by SDS-PAGE followed by immunoblot analyses. ATG13 phosphorylated at S318 was detected only in lanes containing WT ULK1 or the AMPK-resistant mutant (4SA)