Figure 1.
Utilizing CRISPR/Cas9 to Modulate Expression of UTRN, a Disease-Modifying Gene in DMD Myoblasts
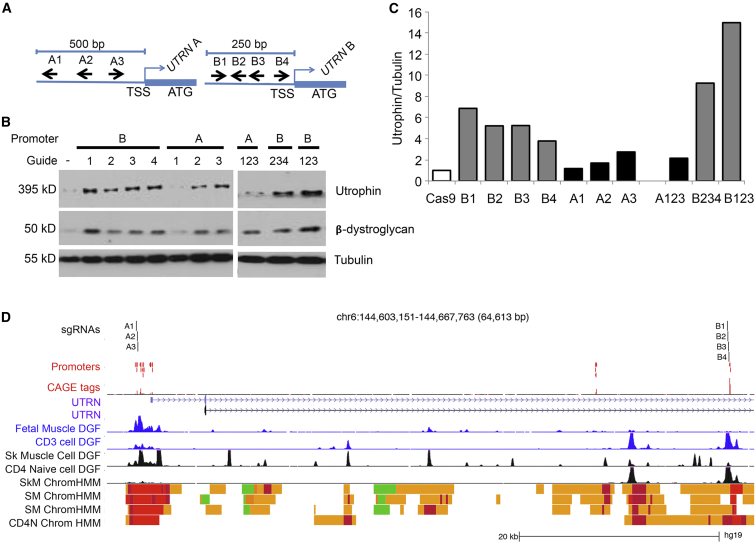
(A) Schematic diagram (not to scale) of sgRNAs targeting regions upstream of UTRN A (A1–A3) and B (B1–B4) TSSs.
(B) CRISPR/Cas9-mediated transcriptional activation of UTRN in DMD myoblasts. Amounts of utrophin, β-dystroglycan, and tubulin were analyzed by western blot 4 days after transfection with dCas9-VP160 plasmid containing each sgRNA.
(C) The amount of utrophin was normalized to that of tubulin by densitometric analysis of four different experiments.
(D) Location of sgRNAs in relation to UTRN TSS, DNase I hypersensitivity footprints, and chromatin-state maps. sgRNAs are plotted above experimentally determined TSSs obtained from a FANTOM5 assay of over 300 primary tissues. The maximum signal at each promoter region is shown below the TSSs (CAGE tags). Digital DNase Footprinting (DGF) assays for fetal muscle and primary CD3 cells are shown in blue (ENCODE). DGF assays for skeletal-muscle cells, skeletal muscle, and naive CD4 cells are shown in black. Chromatin-state maps from the Roadmap Epigenomic Consortium are shown for skeletal-muscle cells (SkM), skeletal muscle (SM), and naive CD4 cells (CD4N). Red indicates TSSs, and yellow indicates enhancer states. The A guides all fall within muscle promoter regions. The B guides fall into an enhancer region immediately upstream of an annotated promoter region. In CD4 cells, this region is considered an active promoter. At promoter B, the DGF footprint in muscle cells is weak in comparison to that in CD4 cells. Data were plotted according to positions from the UCSC Genome Browser. FANTOM5, DGF, and chromatin-state data were obtained from UCSC “Track Hubs.”