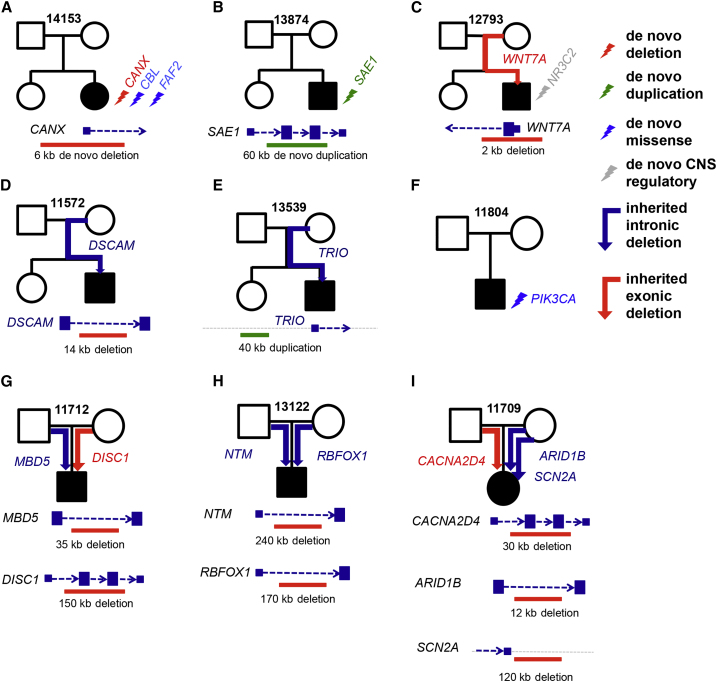
Figure 6.
All Variants Shown for Each Family Were Validated by Appropriate Methods
(A) Family 14153. The events are a de novo exonic deletion of the promoter and first exon of CANX and two de novo missense SNVs in CBL (MIM: 165360) and FAF2. The location of the de novo deletion is also shown with respect to CANX.
(B) Family 13874. The event is a de novo exonic duplication in SAE1. Furthermore, we provided a mock representation of the de novo duplication with respect to SAE1.
(C) Family 12793. The event is a promoter and exonic WNT7A deletion passed from the mother to the male proband. As shown in the mock representation of the inherited deletion, it removes the 5′ UTR and the first exon of WNT7A. This proband and the mother both have macrocephaly, which is in concordance with maternally inherited deletion of WNT7A.
(D) Family 11572. The event is a DSCAM deletion encompassing CNS DNase I hypersensitive sites passed from the mother to the male proband. This individual also suffers from nonfebrile seizures, in concordance with disruption of DSCAM.
(E) Family 13539. The event is a duplication upstream of TRIO. It encompasses CNS DNase I hypersensitive sites and was passed from the mother to the male proband.
(F) Family 11804. A conserved missense de novo mutation (phyloP = 2.57) was found in PIK3CA. This individual has macrocephaly, which is in concordance with disruption of PIK3CA.
(G) Family 11712. We found a maternally inherited rare, private exonic deletion of parts of DISC1 and a 35 kb paternally inherited rare intronic deletion of MBD5. The deletion affecting DISC1 is around 150 kb, deletes a few coding exons of this gene, and is not seen in over 15,000 genotyped control individuals.
(H) Family 13122. We found two large rare deletions that intersect NTM and RBFOX1—one inherited from the father and the other inherited from the mother. The maternally inherited rare deletion is 240 kb and deletes most of the first intron of NTM. This is an extremely rare deletion, given that it was not observed in over 15,000 control individuals. The paternally inherited rare deletion is 170 kb and deletes an intron of RBFOX1.
(I) Family 11709. We found three rare, private deletions affecting genes of interest. First, we found a 30 kb paternally inherited rare, private exonic deletion of CACNA2D4. We also found two maternally inherited rare, private deletions that affect genes of interest. One is a 120 kb exonic rare and private deletion less than 5 kb downstream of SCN2A, and the other is a 12 kb rare deletion of an intron of ARID1B.