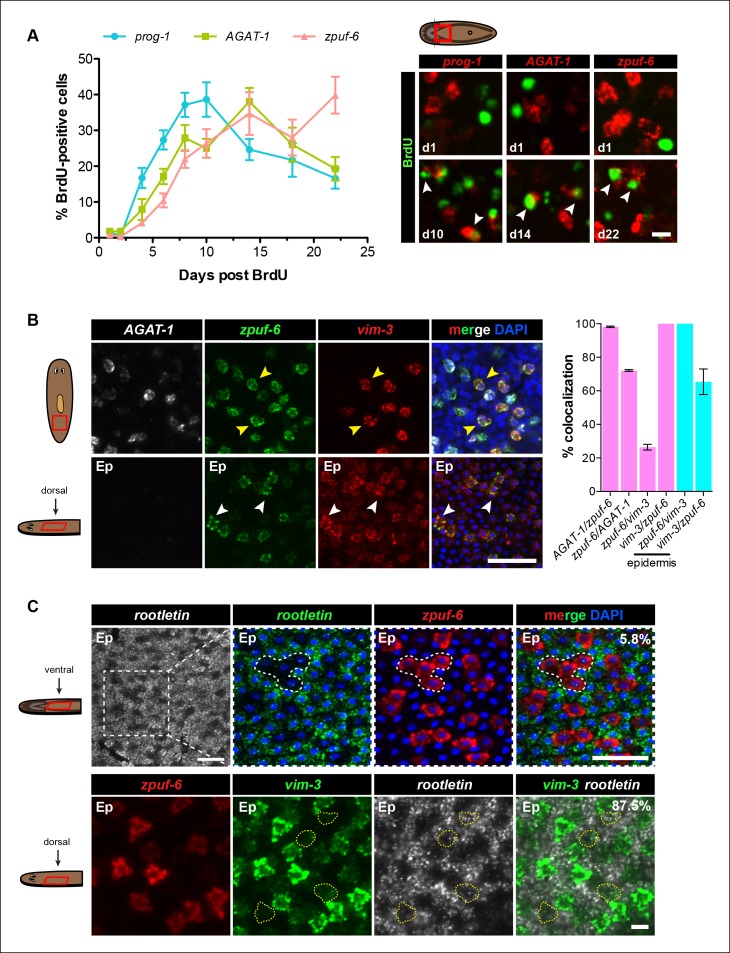
Figure 4. Zpuf-6+ epidermal cells are not terminally differentiated.
(A) Turnover dynamics for prog-1, AGAT-1 and zpuf-6 expressing cell populations. Animals were soaked with BrdU for 24 hr and chased for the indicated time periods. Quantification of the percentage of prog-1+, AGAT-1+ or zpuf-6+ cells analyzed that are BrdU+ are plotted. Error bars: SEM (see Materials and methods). Representative images of BrdU+ cells, are maximum intensity projections over 1 cell diameter of a subset of the quantified images, of the minimum and maximum time points for each gene are shown. White arrowheads highlight double-positive cells. Scale bar: 10 μm. (B) Lineage relationship between AGAT-1, zpuf-6 and vim-3. Top row: Triple FISH showing overlapping expression between AGAT-1, zpuf-6 and vim-3 in the mesenchyme. Yellow arrowheads highlight cells that are zpuf-6+ vim3+ AGAT-1neg. AGAT-1 and vim-3 exhibit little to no co-expression (Figure 2B). Bottom row: Dorsal epidermal (Ep) view of zpuf-6 and vim-3. White arrowheads highlight cells that co-express zpuf-6 and vim-3 but there are additional cells expressing vim-3. Images are single confocal planes. Scale bar: 50 μm. Quantifications of the percent colocalizations of combinations of AGAT-1, zpuf-6 and vim-3 mesenchymal (magenta) and zpuf-6 and vim-3 dorsal epidermal cells (cyan) are shown (∼200-400 cells were quantified over >3 animals; error bars: SD). Gene1/gene2 notation signifies percentage of gene1+ cells that are also positive for gene2 expression. Notable percentages: zpuf-6+/AGAT-1+ (72%), zpuf-6+/vim-3+ (26%). (C) zpuf-6+ epidermal cells express very low levels of rootletin. Top row: double FISH of rootletin and zpuf-6. 5.8%=percentage of strong zpuf-6+ epidermal cells that co-express rootletin (~300 cells). Scale bars: 50 μm. Bottom row: triple FISH of zpuf-6, vim-3 and rootletin in dorsal epidermis. Only overlay of vim-3 and rootletin is shown (far right panel). Yellow-dashed shapes outline cells that express very low levels of vim-3 that also express rootletin. 87.5%=percentage of weak vim-3+ dorsal epidermal cells that co-express rootletin (~200 cells). Images are single confocal planes. Scale bar: 10 μm.