Abstract
AIM: To investigate the association of FAS gene polymorphism with coeliac disease (CD) development.
METHODS: FAS-G670A gene polymorphism, located in a gamma interferon activation site, was studied in 146 unrelated CD patients and 203 healthy ethnically matched controls. The restriction fragment length polymorphism (RFLP) method was used to identify FAS-G670A gene polymorphism.
RESULTS: No significant difference was found in genotype frequency between CD cases and controls. In controls, however, the frequency of the GG genotype was significantly higher in women (26.5%) than in men (12.8%) (OR = 2.44, 95%CI 1.15-5.20, P = 0.020) and it was also higher in men with CD than controls (OR = 2.60, 95%CI 0.96-7.05, P = 0.061). The GG genotype frequency was significantly higher in patients with most severe villous atrophy (Marsh IIIc lesions) (OR = 3.74, 95%CI 1.19-11.82, P = 0.025). A significantly less proportion of men suffered from Marsh IIIc lesions than women (OR = 0.20, 95%CI 0.06-0.68, P = 0.01). The risk of having severe villous atrophy increased with the additive effect of the G allele in women (P = 0.027 for trend, age and gender adjusted).
CONCLUSION: FAS-G670A gene polymorphism is associated with the severity of villous atrophy in CD. Female gender is also associated with the severity of villous atrophy.
INTRODUCTION
The pathogenesis of coeliac disease (CD) is unclear. Although the majority of CD patients are HLA-DQ2 or DQ8 positive, only a small percentage of HLA-DQ2 and DQ8 carriers in the healthy European population develop CD[1]. Human genome screening of CD patients and their families support the polygenic inheritance of the disease[2]. Different genes are involved in the disease susceptibility or in determining clinical course and severity of the lesion[3]. Ciccocioppo et al[4] reported the significant correlation between the degree of villous atrophy (VA) and the level of enterocyte apoptosis. FAS (TNFRSF6) expression increased in the abnormal segment of small intestine in CD[4,5]. Therefore, the epithelial FAS engagement might contribute to the development of villous atrophy[4,5]. We studied the association between the FAS-G670A gene polymorphism with the severity of VA and the disease susceptibility in a cohort of untreated Caucasian CD patients at the time of presentation and ethnically matched healthy controls.
MATERIALS AND METHODS
Subjects
Before treatment, 146 unrelated CD patients were classified according to modified Marsh classification. Their diagnoses were confirmed only if patients responded to gluten-free diet both clinically and histologically[6], and 203 healthy ethnically matched controls were enrolled in this study.
Methods
Villous atrophy classification The histological features were classified according to the modified Marsh classification[7]. In the original classification, Marsh described subtotal villous atrophy as a destructive lesion and called it the Marsh III lesion. In our study, to assess the severity of the histological features, we classified the Marsh III type lesion into 3 subgroups[8]. Briefly, in all subgroups, histological lesions had the significant intraepithelial lymphocytosis (> 30 lymphocytes per 100 epithelial cells). The architectural changes permitted classifying the lesion into three subtypes with increasing severity. They were designated as Marsh IIIa (partial VA) when the villous-crypt ratio was less than 1/1, Marsh IIIb (subtotal VA) when there were still recognizable villi in an otherwise flat mucosa, and Marsh IIIc (total VA) when there was nearly complete absence of villi.
Typing FAS-G670A polymorphism PCR amplification and genotype analysis for FAS-G670A were performed according to previously published methods[9]. In brief, genomic DNA was extracted from peripheral blood using a standard proteinase K digestion and phenol/chloroform extraction method and mouthwash method[10]. The FAS–G670A polymorphism was typed as described previously by Huang et al[9] with the following minor modifications. PCR: 5 min at 95 °C, 30 cycles of 30 s at 95 °C, 30 s at 62 °C, and 1 min at 72 °C, followed by a final extension for 7 min at 72 °C. Primer sequences were 5’-CTA CCT AAG AGC TAT CTA CCG TTC-3’ and 5’-GGC TGT CCA TGT TGT GGC TGC-3’. The 332 bp PCR product was digested with MvaI for 5 h at 37 °C. Allele G yielded three fragments of 99 bp, 189 bp, and 44 bp, whereas allele A yielded two fragments of 99 bp and 233 bp. Digested fragments were separated on 30g/L agarose gels and visualized after ethidium bromide staining (Figure 1).
Figure 1.
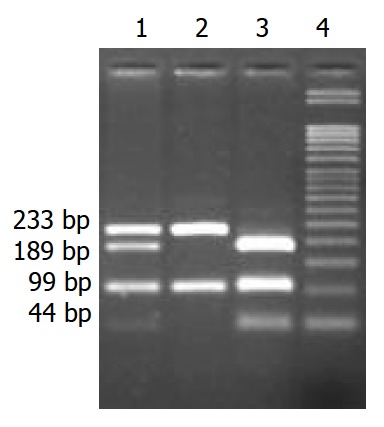
1: GA genotype, 2: AA genotype, 3: GG genotype, 4: 50 bp DNA marker distinguished alleles G (189 bp) and A (233 bp).
Statistical analysis
Hardy–Weinberg equilibrium test was carried out by using statistical software for linkage analysis[11]. Χ 2 statistics and Fisher’s exact test were used for comparisons of frequencies. The subjects were classified according to the number of G alleles that they inherited, such as 0 for subjects who were carriers for A allele of FAS-G670A, i.e. AA genotype, 1 for those who were carriers of 1 copy of G allele, i.e. AG genotype and 2 for subjects who inherited 2 copies of G allele, i.e. GG genotype. Logistic regression was used to fit statistical models to predict the association of FAS-G670A polymorphism with the severity of villous atrophy or with susceptibility to CD. All the statistical models were adjusted for age (years) and gender. Associations are expressed as odd ratios (OR) with 95% confidence interval (95%CI). Estimation of 95%CI was based on Wald’s method. A two tailed P value < 0.05 was considered as significant. Statistical analysis was performed with SPSS version 10.07 for windows software.
RESULTS
Susceptibility of CD
The characteristics of study participants are shown in Table 1. The genotype frequencies were in Hardy–Weinberg equilibrium proportions in patients and controls. There was no significant difference in genotype distributions between patients and controls (Table 1). In men, the frequency of GG genotype was higher in cases (27.3%) compared to that of controls (12.8%), yielding to OR of 2.60, 95%CI 0.96-7.05, P = 0.061. The GG genotype was significantly (P = 0.020) higher in women (26.5%) compared with men (12.8%) in healthy control.
Table 1.
Characteristics and genotype frequencies of FAS-G670A in untreated CD with villous atrophy
| CD (n = 146) | Controls (n = 203) | OR1 (95%CI) | OR2 (95%CI) | OR3 (95%CI) | |
| Age (mean ± SD) yr. | 38.7 ± 20.0 | 39.1 ± 11.9 | |||
| Gender (F/M) | 113/33 | 117/86 | |||
| GG genotype (F/M) | 25/9 | 31/11 | 1.08 (0.64-1.82) | 2.44 (1.15-5.20) | 0.75 (0.31-1.82) |
| GA genotype (F/M) | 60/15 | 66/49 | 0.79 (0.51-1.22) | 0.99 (0.57-1.75) | 1.37 (0.63-2.99) |
| AA genotype(F/M) | 28/9 | 20/26 | 1.30 (0.78-2.16) | 0.46 (0.24-0.91) | 0.88 (0.37-2.11) |
F, women; M, men, OR1, age and gender adjusted, CD vs controls, OR2, age adjusted, F vs M in healthy controls, OR3, age adjusted, F vs M in CD.
Association of villous atrophy
In cases, men had a significantly lower frequency of Marsh IIIc compared to women (OR = 0.20, 95%CI 0.06-0.68, P = 0.01). Genotype and allele frequencies in Marsh III subgroups are presented in Table 2. There was a significant (P = 0.025) difference of GG genotype frequency between Marsh IIIc which had the most severe form of the disease and Marsh IIIa (OR = 3.74, 95%CI 1.19-11.82). In chromosomal analysis, we found a borderline significant association between G allele and Marsh IIIc villous atrophy (OR = 1.81, 95%CI 0.97-3.38, Table 2). However, in women, G allele was significantly associated with this subgroup (OR = 1.75, 95%CI 1.00-3.06, P = 0.048). The severity of VA increased additively with increasing number of G alleles in women (Pfor trend = 0.027) (Figure 2).
Table 2.
Association of severity of villous atrophy with FAS-G670A polymorphism in untreated CD
| Marsh |
Genotype analysis |
Allelic analysis |
|||||
| GG genotype | GA genotype | AA genotype | OR1 (95%CI) | G allele (%) | A allele (%) | OR2 (95%CI) | |
| F/M | F/M | F/M | |||||
| IIIa (%) | 3/3 (15.0) | 17/6 (57.5) | 6/5 (27.5) | Reference | 35 (44.0) | 45 (56.0) | Reference |
| IIIb (%) | 9/5 (21.5) | 25/7 (49.2) | 15/4 (29.2) | 1.63 (0.56-4.76) | 60 (46.0) | 70 (54.0) | 1.10 (0.63-1.93) |
| IIIc (%) | 13/1 (34.1)3 | 18/2 (48.8) | 7/0 (17.1) | 3.74 (1.19-11.82) | 48 (58.5) | 34 (41.5) | 1.81 (0.97-3.38) |
F, women; M, men, OR1, adjusted, for GG genotype frequency;
P = 0.025, OR2, adjusted, for G allele frequency.
Figure 2.
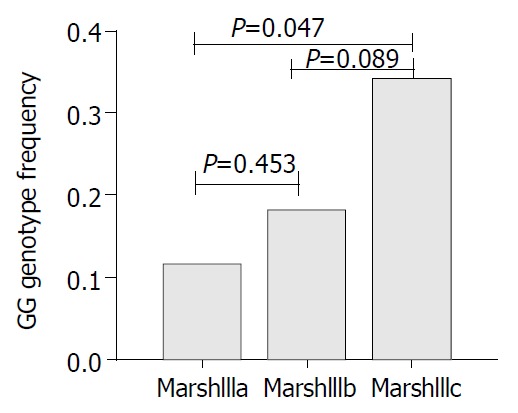
Trend for GG genotype frequency in different severity of villous atrophy in untreated CD women, P = 0.027, adjusted for age.
DISCUSSION
We found a significant association between the FAS–670 GG genotype and the severity of celiac disease, particularly in women. However, we did not find significant differences in genotype frequencies between controls and CD patients. This suggested that the FAS gene was not a susceptible gene for this disease. The fact that genotype frequencies of patients and controls were in Hardy Weinberg equilibrium confirmed the validity of the typing of the results. The intestinal pathological diagnosis was confirmed by two independent pathologists. Therefore our data suggested an association of FAS-G670A polymorphism with the severity of CD.
Since this polymorphism of the FAS gene (TNFRSF6) is located in the promoter region, it may affect the level of transcription of the FAS protein. Previous work suggested that the substitution of G to A in the position –670 (TTCCAG G/A AA) would change the gamma interferon activation site (GAS) (TTCnnnGAA)[12-14]. This site was involved in interferon gamma and interferon alpha signalling[15]. GAS elements are known to bind to homodimers of a phosphorylated form of the 91-kDa transcription factor, STAT1. Interferon gamma could cause tyrosine phosphorylation of STAT1 by the interferon gamma receptor-associated Janus kinases 1 and 2. Subsequently, phosphorylated STAT1 formed homodimers and translocated into the nucleus where it induced transcription of GAS-containing genes[16]. FAS was significantly upregulated by interferon gamma according to several reports[17-19]. Xu et al[20] and De Saint Jean et al[21] have implicated STAT1 in this upregulation effect of interferon gamma. We therefore postulate that FAS-670G variant containing the GAS (TTCCAGGAA) could be affected by interferon gamma production and increase the transcription of Fas. This may result in different degree of apoptosis in CD with different degrees of VA. Our data confirmed that the risk of having severe villous atrophy increased additively with the number of G alleles. The GG homozygote was strongly correlated to the severity of VA in CD. Intermediated by GAS, both interferon gamma and Fas might play an important role in the pathogenesis of villous atrophy. This was in agreement with previous reports showing that mucosal gluten exposure elicited a high level interferon gamma expression in CD[22]. Interferon gamma could increase FAS-induced apoptosis of human intestinal epithelial cells in a dose-dependent manner[23].
Considering the previous reports[24-26], we hypothesize that FAS-G670A is functional and further functional tests are necessary. The FAS-G670A gene polymorphism contributes to the determination of the severity of small intestinal lesions and opens a new area of research that may help understand the heterogeneity of this disease. Our findings were in the same line of a recent genome-wide family-based linkage study of CD that found a potential susceptible locus at 10q23.1[27] close to the cytogenetic region (10q23.31) of the FAS (TNFRSF6) gene[28]. These results may also explain why the different genome-wide studies of families with multiple cases of CD had not uniformly found a similar lod score at chromosome 10q23, since the proportion of patients with severe villous atrophy might differ from each other in those studies.
Our data also showed a gender difference in relation to the severity of VA and in the genotype distribution of FAS-G670A. Even though there was no difference of GG genotype frequency in women and men among the cases, men had a significantly less proportion of Marsh IIIc VA than women. This might be due to the different immune responses between men and women. Women were more likely to develop Th1 response (secreting higher amounts of IL-2, interferon gamma, and TNF-beta than men)[29]. In summary, our findings suggest that increased expression of FAS in villous atrophy is in part genetically regulated and the FAS gene plays a significant role.
ACKNOWLEDGEMENTS
The authors would like to express thanks to JBA Crusius PhD, A. Zwiers MSc and Ms. M Demirkaya for their laboratory support, and Ms. L Atjak for her secretarial assistance.
Footnotes
Supported by the Chinese Scholarship Council, No. 98932034
Edited by Gupat MK and Wang XL
References
- 1.Schuppan D. Current concepts of celiac disease pathogenesis. Gastroenterology. 2000;119:234–242. doi: 10.1053/gast.2000.8521. [DOI] [PubMed] [Google Scholar]
- 2.King AL, Ciclitira PJ. Celiac disease: strongly heritable, oligogenic, but genetically complex. Mol Genet Metab. 2000;71:70–75. doi: 10.1006/mgme.2000.3067. [DOI] [PubMed] [Google Scholar]
- 3.Fraser JS, Ciclitira PJ. Pathogenesis of coeliac disease: implications for treatment. World J Gastroenterol. 2001;7:772–776. doi: 10.3748/wjg.v7.i6.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciccocioppo R, Di Sabatino A, Parroni R, Muzi P, D'Alò S, Ventura T, Pistoia MA, Cifone MG, Corazza GR. Increased enterocyte apoptosis and Fas-Fas ligand system in celiac disease. Am J Clin Pathol. 2001;115:494–503. doi: 10.1309/UV54-BHP3-A66B-0QUD. [DOI] [PubMed] [Google Scholar]
- 5.Maiuri L, Ciacci C, Raia V, Vacca L, Ricciardelli I, Raimondi F, Auricchio S, Quaratino S, Londei M. FAS engagement drives apoptosis of enterocytes of coeliac patients. Gut. 2001;48:418–424. doi: 10.1136/gut.48.3.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Revised criteria for diagnosis of coeliac disease. Report of Working Group of European Society of Paediatric Gastroenterology and Nutrition. Arch Dis Child. 1990;65:909–911. doi: 10.1136/adc.65.8.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marsh MN. Gluten, major histocompatibility complex, and the small intestine. A molecular and immunobiologic approach to the spectrum of gluten sensitivity ('celiac sprue') Gastroenterology. 1992;102:330–354. [PubMed] [Google Scholar]
- 8.Rostami K, Kerckhaert J, Tiemessen R, von Blomberg BM, Meijer JW, Mulder CJ. Sensitivity of antiendomysium and antigliadin antibodies in untreated celiac disease: disappointing in clinical practice. Am J Gastroenterol. 1999;94:888–894. doi: 10.1111/j.1572-0241.1999.983_f.x. [DOI] [PubMed] [Google Scholar]
- 9.Huang QR, Morris D, Manolios N. Identification and characterization of polymorphisms in the promoter region of the human Apo-1/Fas (CD95) gene. Mol Immunol. 1997;34:577–582. doi: 10.1016/s0161-5890(97)00081-3. [DOI] [PubMed] [Google Scholar]
- 10.Laine ML, Farré MA, Crusius JB, van Winkelhoff AJ, Peña AS. The mouthwash: a non-invasive sampling method to study cytokine gene polymorphisms. J Periodontol. 2000;71:1315–1318. doi: 10.1902/jop.2000.71.8.1315. [DOI] [PubMed] [Google Scholar]
- 11.Ott J. Utility programs for analyisis of genetic linkage; Program HWE; 1988; URL: http: //www.hgmp.mrc.ac.uk/Registered/ Help/linkutil/ [Google Scholar]
- 12.Decker T, Kovarik P, Meinke A. GAS elements: a few nucleotides with a major impact on cytokine-induced gene expression. J Interferon Cytokine Res. 1997;17:121–134. doi: 10.1089/jir.1997.17.121. [DOI] [PubMed] [Google Scholar]
- 13.Chatterjee-Kishore M, Wright KL, Ting JP, Stark GR. How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J. 2000;19:4111–4122. doi: 10.1093/emboj/19.15.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bauvois B, Djavaheri-Mergny M, Rouillard D, Dumont J, Wietzerbin J. Regulation of CD26/DPPIV gene expression by interferons and retinoic acid in tumor B cells. Oncogene. 2000;19:265–272. doi: 10.1038/sj.onc.1203292. [DOI] [PubMed] [Google Scholar]
- 15.Shuai K. Interferon-activated signal transduction to the nucleus. Curr Opin Cell Biol. 1994;6:253–259. doi: 10.1016/0955-0674(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 16.Gao J, Morrison DC, Parmely TJ, Russell SW, Murphy WJ. An interferon-gamma-activated site (GAS) is necessary for full expression of the mouse iNOS gene in response to interferon-gamma and lipopolysaccharide. J Biol Chem. 1997;272:1226–1230. doi: 10.1074/jbc.272.2.1226. [DOI] [PubMed] [Google Scholar]
- 17.Schwartzberg LS, Petak I, Stewart C, Turner PK, Ashley J, Tillman DM, Douglas L, Tan M, Billups C, Mihalik R, et al. Modulation of the Fas signaling pathway by IFN-gamma in therapy of colon cancer: phase I trial and correlative studies of IFN-gamma, 5-fluorouracil, and leucovorin. Clin Cancer Res. 2002;8:2488–2498. [PubMed] [Google Scholar]
- 18.Dai C, Krantz SB. Interferon gamma induces upregulation and activation of caspases 1, 3, and 8 to produce apoptosis in human erythroid progenitor cells. Blood. 1999;93:3309–3316. [PubMed] [Google Scholar]
- 19.Pouly S, Becher B, Blain M, Antel JP. Interferon-gamma modulates human oligodendrocyte susceptibility to Fas-mediated apoptosis. J Neuropathol Exp Neurol. 2000;59:280–286. doi: 10.1093/jnen/59.4.280. [DOI] [PubMed] [Google Scholar]
- 20.Xu X, Fu XY, Plate J, Chong AS. IFN-gamma induces cell growth inhibition by Fas-mediated apoptosis: requirement of STAT1 protein for up-regulation of Fas and FasL expression. Cancer Res. 1998;58:2832–2837. [PubMed] [Google Scholar]
- 21.De Saint Jean M, Brignole F, Feldmann G, Goguel A, Baudouin C. Interferon-gamma induces apoptosis and expression of inflammation-related proteins in Chang conjunctival cells. Invest Ophthalmol Vis Sci. 1999;40:2199–2212. [PubMed] [Google Scholar]
- 22.Nilsen EM, Jahnsen FL, Lundin KE, Johansen FE, Fausa O, Sollid LM, Jahnsen J, Scott H, Brandtzaeg P. Gluten induces an intestinal cytokine response strongly dominated by interferon gamma in patients with celiac disease. Gastroenterology. 1998;115:551–563. doi: 10.1016/s0016-5085(98)70134-9. [DOI] [PubMed] [Google Scholar]
- 23.Ruemmele FM, Russo P, Beaulieu J, Dionne S, Levy E, Lentze MJ, Seidman EG. Susceptibility to FAS-induced apoptosis in human nontumoral enterocytes: role of costimulatory factors. J Cell Physiol. 1999;181:45–54. doi: 10.1002/(SICI)1097-4652(199910)181:1<45::AID-JCP5>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 24.Huang QR, Danis V, Lassere M, Edmonds J, Manolios N. Evaluation of a new Apo-1/Fas promoter polymorphism in rheumatoid arthritis and systemic lupus erythematosus patients. Rheumatology (Oxford) 1999;38:645–651. doi: 10.1093/rheumatology/38.7.645. [DOI] [PubMed] [Google Scholar]
- 25.Feuk L, Prince JA, Breen G, Emahazion T, Carothers A, St Clair D, Brookes AJ. apolipoprotein-E dependent role for the FAS receptor in early onset Alzheimer's disease: finding of a positive association for a polymorphism in the TNFRSF6 gene. Hum Genet. 2000;107:391–396. doi: 10.1007/s004390000383. [DOI] [PubMed] [Google Scholar]
- 26.Bolstad AI, Wargelius A, Nakken B, Haga HJ, Jonsson R. Fas and Fas ligand gene polymorphisms in primary Sjögren's syndrome. J Rheumatol. 2000;27:2397–2405. [PubMed] [Google Scholar]
- 27.King AL, Yiannakou JY, Brett PM, Curtis D, Morris MA, Dearlove AM, Rhodes M, Rosen-Bronson S, Mathew C, Ellis HJ, et al. A genome-wide family-based linkage study of coeliac disease. Ann Hum Genet. 2000;64:479–490. doi: 10.1046/j.1469-1809.2000.6460479.x. [DOI] [PubMed] [Google Scholar]
- 28.http://genome.ucsc.edu/, Assembly Human April 2003 [Google Scholar]
- 29.Whitacre CC, Reingold SC, O'Looney PA. A gender gap in autoimmunity. Science. 1999;283:1277–1278. doi: 10.1126/science.283.5406.1277. [DOI] [PubMed] [Google Scholar]


