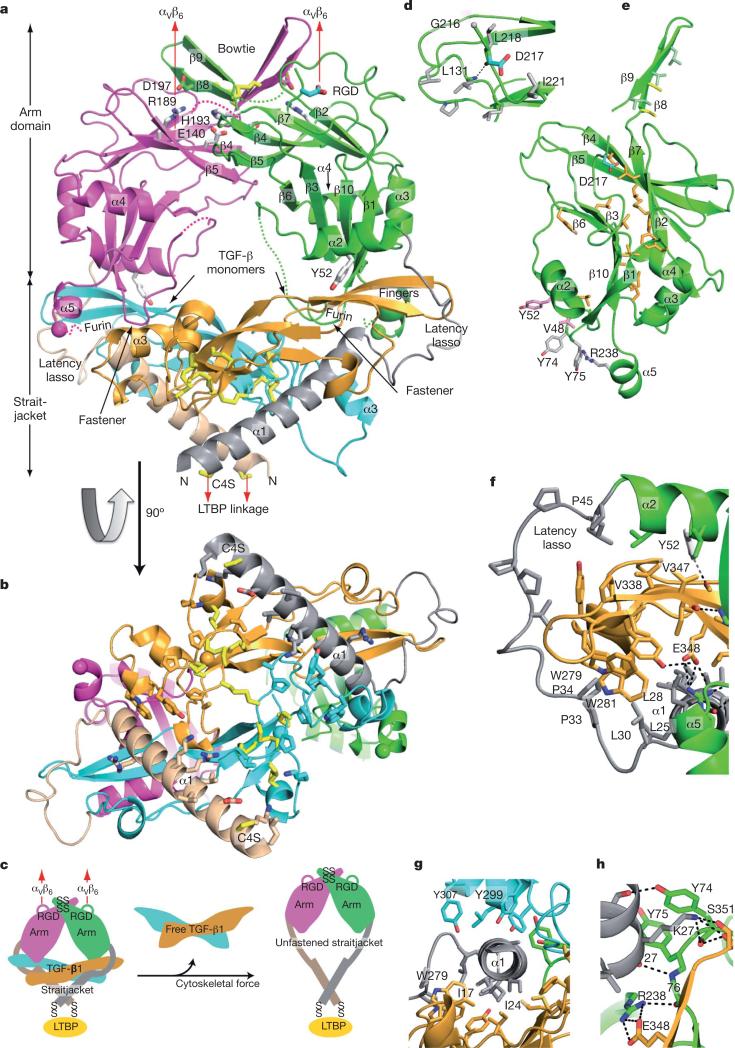
Figure 1. Architecture of proTGF-β1.
Arm, straitjacket and TGF-β1 monomer segments are coloured differently. a, b, Overall structure. Spheres mark the last residue visible in density in the prodomain and the first residue of the growth factor. Disordered segments are dashed. Red arrows show the directions of forces during activation by integrins. Key side chains are shown in stick representation, including Asp of the RGD motif in cyan and CED mutations in white. Disulphide bonds and the Cys 4 mutation to Ser are in yellow. c, Schematic of the structure and activation mechanism. SS, disulphide bonds. d, Hydrophobic residues near Asp 217 of the RGD motif. e, Arm domain. Side chains for the hydrophobic core are shown in gold (also marked in Fig. 2 and Supplementary Fig. 2), conserved α2-helix residues that interact with the growth factor are in pink, fastener residues are in silver and bowtie residues are in light green for aliphatics and yellow for Cys. f–h, Straitjacket and fastener details.