Figure 2.
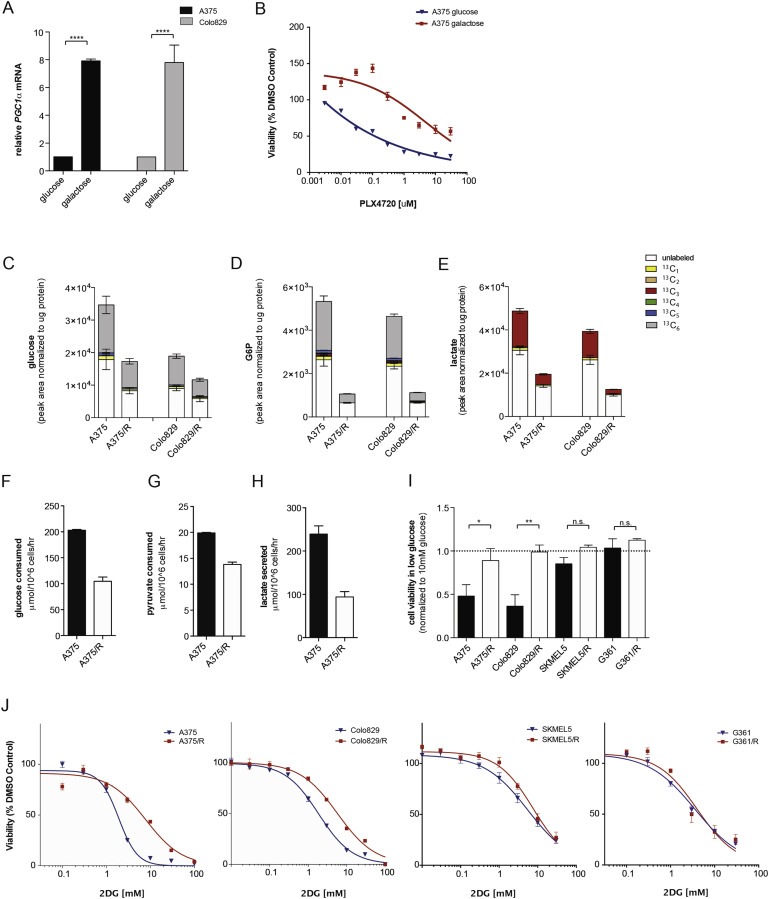
BRAF inhibitor resistant melanoma cells display decreased glycolytic flux. (A) Graph showing PGC1α mRNA levels in A375 and Colo829 cells grown for 5 days in medium containing glucose or galactose as the sole carbon source. Error bars represent SEM of at least two independent experiments with 3 biological replicates. ****p < 0.0001. (B) Graph showing A375 cell growth (sulforhodamine B) in glucose or galactose containing medium in the presence of increasing concentrations of PLX4720 for 72 h. (C–E) Graphs showing glucose (C), glucose‐6‐phosphate (D), and lactate (E) levels in A375, Colo829, A375/R and Colo829/R cells labeled with U‐13C6 glucose. The graphs show mean ± SD (n = 6) and similar results were observed in two independent experiments. Legend displays the different isotopomers. 13C1: one carbon atom labeled; 13C2 – 13C6: two to six carbon atoms labeled. Glucose and glucose‐6‐phosphate are 6‐carbon molecules hence 13C6 labeling (gray). Lactate is a 3‐carbon molecule hence the 13C3 labeling (red). Endogenous metabolites are unlabeled and shown in white. (F–H) Graphs showing glucose (F), pyruvate, (G) and lactate (H) consumption (extracellular exchange rate) in A375 and A375/R cells. Data are representative of three independent studies and displayed as mean ± SD of n = 6. (I) Growth (sulforhodamine B) of A375, A375/R, Colo829, Colo829/R, SKMEL5, SKMEL5/R, G361 and G361/R cells in medium containing 1 mM glucose for 72 h. Data are expressed relative to growth in medium containing 10 mM glucose (dotted line). Error bars represent SEM of at three independent experiments with 3 biological replicates. *p < 0.05, **p < 0.01, n.s.: non‐significant. (J) Growth of A375, A375/R, Colo829, Colo829/R, SKMEL5, SKMEL5/R, G361 and G361/R cells in the presence of increasing concentrations 2DG for 72 h (n = 3).
