Figure 1. Local chromatin remodelling by pioneers.
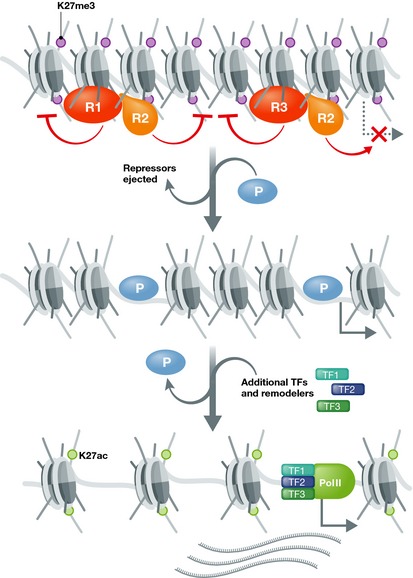
A genomic region is condensed, and genes within are repressed. Repressor proteins (R1‐3) such as Mbd3, Utf1 and Tbx3 maintain the repressive chromatin state and reinforce transcriptional silence. Once available, pioneer factor complexes (P) like NeuroD1 + partners find and bind their target sites, including sites within repressive chromatin. This then recruits enzymatic activities to modulate repression locally. For example, repressor proteins like Tbx3 and Mbd3 are ejected and histone trimethylation of H3K27 is exchanged for acetylation. Chromatin locally decondenses in the process and “opens” for targeting by settler TFs (TF1‐3) to regulate gene expression either in conjunction with, or independent of the pioneer complex. Note that each nucleosome contains two H3K27 positions and is modified on a multitude of histone tail residues; for simplicity, only trimethylation or acetylation on H3K27 is shown. Absence of modifications in the middle panel is meant to indicate uncertainty regarding the exchange kinetics. Complex context‐dependent regulation is achieved by, for example, (i) availability of pioneer binding partners / cofactors, (ii) aspects of the chromatin environment, including recruiting factors already present, (iii) the mode of pioneer interaction with the chromatin (e.g. displacement of proteins, delivery of HAT activities, etc.) and (iv) the specific availability of TFs capable of translating the regulatory information encoded in open chromatin into local gene activity.
