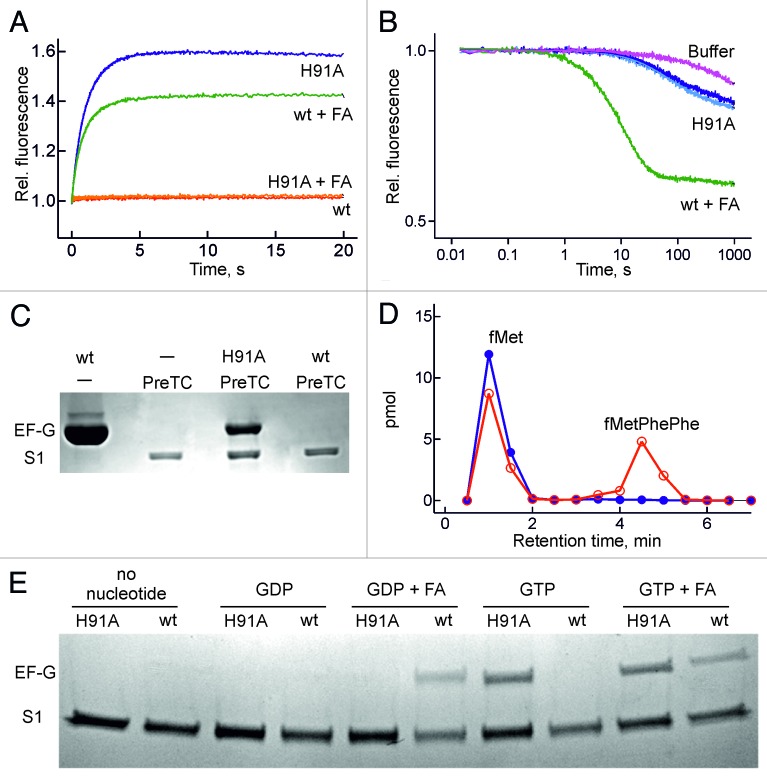
Figure 4. Formation of a stable ribosome-EF-G(H91A) -GTP complex. (A) Time courses monitored by mant-nucleotide fluorescence. Red trace, wt EF-G + mant-GTP; green trace, wt EF-G with mant-GDP and FA (0.2 mM); blue trace, EF-G(H91A) + mant-GTP; orange trace, EF-G(H91A) + mant-GDP with FA. (B) Stability of ribosome-EF-G-mant-GTP complexes. The complex with wt EF-G + FA was chased with excess GDP (green trace), the complex with EF-G(H91A) with excess GDP (dark blue trace) or GTP (light blue trace). (C) EF-G binding to pre-translocation complex (PreTC). EF-G and ribosomal protein S1 were visualized by Coomassie staining. PreTC, pre-translocation complex. (D) Peptide formation. Initiation complexes programed with MFF-mRNA and containing fMet-tRNAfMet in the P site were pre-incubated with wt EF-G (blue) or EF-G(H91A) (red) in the presence of GTP and mixed with EF-Tu–GTP-Phe-tRNAPhe. fMetPhePhe tripeptide was separated from unreacted fMet by HPLC (Materials and methods). (E) eF-G binding to vacant ribosomes. conditions as in (C) but with 5 µM mant-nucleotides and 0.2 mM FA for comparison with Fig. 4A and 4B.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.