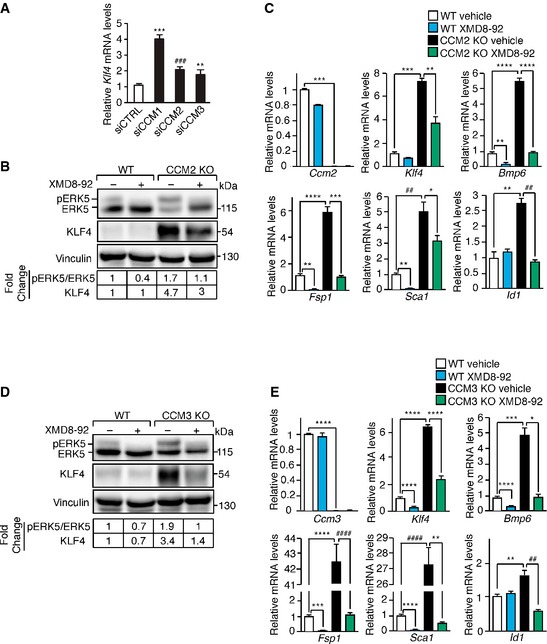
qRT–PCR analysis of Klf4 in WT ECs transfected with either siRNA directed to anyone of the three Ccm genes or control siRNA. Data are presented as mean ± SD (n = 3). Results are shown as fold changes relative to control siRNA‐treated ECs. A two‐tailed unpaired t‐test was performed. ***P = 0.0001, ###
P = 0.0009, **P = 0.0017.
WB analysis of pERK5, ERK5, and KLF4 in lung‐derived WT and CCM2 KO ECs treated with XMD8‐92 or vehicle for 72 h. Both pERK5/ERK5 ratio and KLF4 amount normalized over vinculin, the loading control, were quantified by densitometry scan. WB results are representative of three independent observations.
qRT–PCR of Ccm2, Klf4, Bmp6, and some EndMT markers in WT and CCM2 KO ECs treated with XMD8‐92 or vehicle for 72 h. qRT–PCR results are shown as mean ± SD (n = 3), and fold changes are relative to vehicle‐treated WT ECs. A two‐tailed unpaired t‐test was performed. Ccm2: ***P = 0.0003; Klf4: ***P = 0.0002, **P = 0.006; Bmp6: **P = 0.0018, ****P = 8.7E‐05; Fsp1: **P = 0.0013, ****P = 6.5E‐05, ***P = 0.0008; Sca1: **P = 0.001, ##
P = 0.0025, *P = 0.029; Id1: **P = 0.0013, ##
P = 0.0029.
WB analysis of pERK5, ERK5, and KLF4 in lung‐derived WT and CCM3 KO ECs treated with XMD8‐92 or vehicle for 72 h. Both pERK5/ERK5 ratio and KLF4 were normalized and quantified as in (A).
qRT–PCR of Ccm3, Klf4, Bmp6, and some EndMT markers in WT and CCM3 KO ECs treated with XMD8‐92 or vehicle for 72 h quantified as in (C). Ccm3: ****P < 0.00001; Klf4: ****P < 0.00001; Bmp6: ****P < 0.00001, ***P = 0.0009, *P = 0.01; Fsp1: ***P = 0.0002, ****P < 0.00001, ####
P = 3.8E‐05; Sca1: ****P < 0.00001, ####
P = 4.79E‐05, **P = 0.0013; Id1: **P = 0.0039, ##
P = 0.0017.
Source data are available online for this figure.