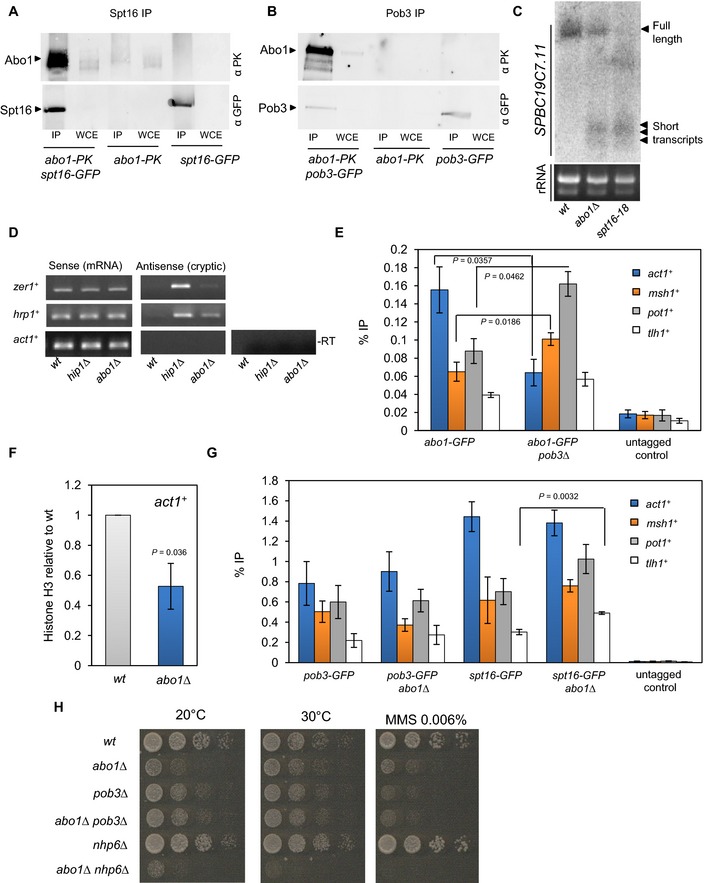
Figure 3. Abo1 associates with FACT and suppresses cryptic transcription.
-
A, BWhole‐cell extracts (WCE) were prepared from the indicated strains, immunoprecipitated (IP) with anti‐GFP antibody and subjected to Western blotting with anti‐V5‐Pk (Serotec) and anti‐GFP (Life Technologies) antibodies. Data are representative of three independent biological repeats.
-
CRNA purified from wild‐type, abo1∆ and spt16‐18 cells was analysed by Northern blotting using a probe to the 3′ end of SPBC19C7.11 (top panel). RNA (5 μg) used for Northern blotting was analysed on an ethidium bromide‐stained 1% TAE agarose gel (bottom panel). Data are representative of two independent biological repeats.
-
DRNA purified from wild‐type and abo1∆ cells was analysed by strand‐specific RT–PCR. RNA from hip1∆ cells was analysed as a control. One primer, complementary to either the forward or reverse transcripts, was included during the reverse transcription step, and the second primer was then added during PCR amplification. Control reactions omitting the reverse transcription step (−RT) were included to demonstrate the absence of contaminating genomic DNA. Data are representative of two independent biological repeats.
-
EThe indicated strains were subjected to ChIP analysis with anti‐GFP antibodies, and the resulting DNA was analysed by qPCR for the indicated locus. Data are the mean of three independent biological repeats, and error bars represent ± SEM. P‐values were calculated using a two‐tailed unpaired t‐test. Significant (P < 0.05) differences are indicated.
-
FHistone H3 levels at act1 + were determined by ChIP–qPCR. Data are the mean of three independent biological repeats, and error bars represent ± SEM. P‐value was calculated using a two‐tailed unpaired t‐test.
-
GThe indicated strains were subjected to ChIP analysis as described for (E).
-
HLog‐phase cells were subjected to five‐fold serial dilution and spotted onto rich (YE5S) agar or agar supplemented with MMS (0.006%). Plates were incubated for 3–5 days at 30°C or 7 days at 20°C. Images are representative of three independent biological repeats.
Source data are available online for this figure.
