Figure 7. Abo1 is necessary for accurate chromosome segregation.
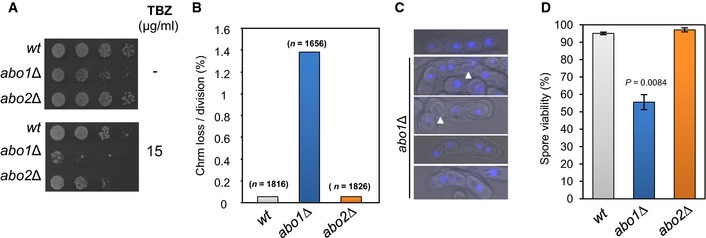
- Log‐phase cells were subjected to five‐fold serial dilution and spotted onto rich (YE5S) agar or agar supplemented with TBZ (15 μg/ml) and incubated for 3–5 days at 30°C. Data are representative of three independent biological repeats.
- The frequency of Ch16 minichromosome loss was assayed by plating cells onto adenine‐limiting agar and determining the frequency of half‐sectored (red‐white) colonies. At least two independently derived strain isolates were used for each genotype, and the total number of colonies counted for each genotype is indicated in parentheses.
- Heterothallic (h 90) strains were cultured on nitrogen‐limiting (EMMG) medium for 3 days at 25°C to induce mating and meiosis. The resulting asci were stained with DAPI and visualised by fluorescence microscopy. Spores lacking a DAPI signal are indicated by arrowheads. Data are representative of three biological repeats.
- Spore viability was measured by dissecting spores onto YE5S agar followed by incubation for 4–5 days at 30°C. Data are the mean of three independent biological repeats, and error bars represent ± SEM. P‐value was calculated using a two‐tailed unpaired t‐test.
