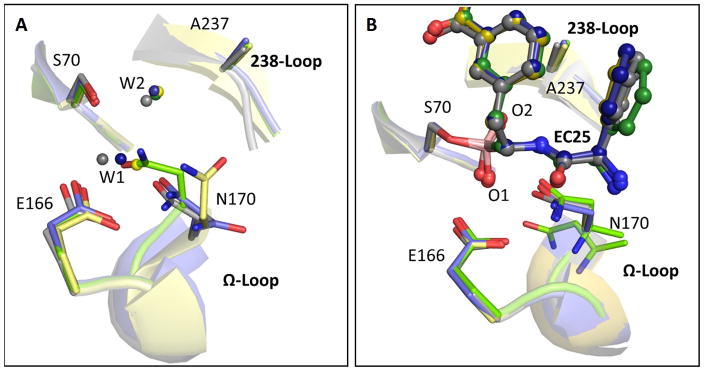
Fig. 4. The mutations impair TEM-1’s catalytic pre-organization.
Superposition of the key active-sites residues in wild-type (gray), the v13 single G238S (blue) and R164S (yellow) mutants, and the v13 double mutant R164S/G238S (green). (A) Ligand-free structures. (B) The EC25 inhibitor complexes (EC25 is shown in ball-sticks). In the free structures, W1 represents the de-acylating water and W2 in the oxyanion-hole water44. In the EC25-bound structures, the borate’s oxygen atoms, marked as O1 and O2, sit in the locations of W1 and W2, respectively. Note the dual conformation of N170 in the EC25 complex of R164S/G238S.