SUMMARY
Introducing a carbon concentrating mechanism and a faster Rubisco from cyanobacteria into higher plant chloroplasts could improve photosynthetic performance by increasing the rate of CO2 fixation while decreasing losses caused by photorespiration. We previously demonstrated that tobacco plants will grow photoautotrophically using Synechococcus elongatus Rubisco, although the plants exhibited considerably slower growth than wild-type and required supplementary CO2. Because of concerns that vascular plant assembly factors might not be adequate for assembly of a cyanobacterial Rubisco, prior transgenic plants included the cyanobacterial chaperone RbcX or the carboxysomal protein CcmM35. Here we show that neither RbcX nor CcmM35 is needed for assembly of active cyanobacterial Rubisco. Furthermore, by altering the gene regulatory sequences on the Rubisco transgenes, cyanobacterial Rubisco expression was enhanced and the transgenic plants grew at near wild-type growth rates, though still requiring elevated CO2. We performed detailed kinetic characterization of the enzymes produced with and without the RbcX and CcmM35 cyanobacterial proteins. These transgenic plants exhibit photosynthetic characteristics that confirm the predicted benefits of non-native forms of Rubisco with higher carboxylation rate constants in vascular plants and the potential nitrogen use efficiency that may be gained provided that adequate CO2 can be concentrated near the enzyme.
Keywords: Rubisco, photosynthesis, CO2 concentration mechanism, chloroplast transformation, Synechococcus elongatus, Nicotiana tabacum, KT203393, KM102745, KM102746, KT203394
INTRODUCTION
In photosynthetic organisms, Rubisco (D-ribulose-1,5-bisphosphate carboxylase/oxygenase) catalyses the assimilation of atmospheric carbon dioxide (CO2) into the organic carbon of plant biomass (Andersson and Backlund, 2008; Tabita et al., 2008). Rubisco also catalyses a competing and wasteful reaction with oxygen that initiates photorespiration and the consequent release of previously fixed CO2, NH3 and energy (Parry et al., 2003). Furthermore, because Rubisco is slow, very large amounts are needed (up to 50% of leaf soluble protein) to support adequate photosynthetic rates. Overcoming these limitations of Rubisco is a widely recognized target for increasing photosynthesis and yield (Parry et al., 2003; Whitney et al., 2011; Ort et al., 2015). Furthermore, being able to use less Rubisco would reduce the demand for leaf nitrogen. For example, experiments with wheat have demonstrated that 25% of leaf nitrogen is invested in Rubisco (Evans, 1989), while cyanobacteria Rubisco consumes only 3–5% of nitrogen in cyanobacteria (Price et al., 2013).
Various strategies, generically termed carbon concentrating mechanisms (CCMs), have evolved in nature to concentrate CO2 in the vicinity of Rubisco. In the cyanobacterial CCM, Rubisco is packaged within proteinaceous micro-compartments called carboxysomes (Bobik, 2006; Cheng et al., 2008). The presence of a CCM allows cyanobacteria to use faster forms of Rubisco, albeit with lower affinities for CO2 (the and for Synechococcus PCC6301 Rubisco and tobacco Rubisco are ~12 s−1, ~200 μM and ~3 s−1, ~10.7 μM, respectively (Mueller-Cajar and Whitney, 2008; Whitney et al., 2011)). Introducing such CCMs into crop plants is an attractive target for increasing photosynthetic performance, nitrogen use efficiency and yield (Price et al., 2013; Zarzycki et al., 2013; McGrath and Long, 2014). Furthermore, this mechanism operates on an intracellular level, obviating the need for the gross anatomical requirements associated with the CCM of C4 plants.
Assembly of Rubisco in cyanobacteria requires requires the chaperonin GroEL and its cofactor GroES, which have homologs in chloroplasts named Cpn60, Cpn10 and Cpn20 (Vitlin Gruber et al., 2013). RbcX dimers have been shown to play an essential role in stabilizing the LSU dimers of cyanobacterial Rubisco during its assembly into a core complex and functional holoenzyme, stabilizing the LSU core immediately prior to SSU integration (Liu et al., 2010). However, certain cyanobacterial strains such as S. elongatus PC7942 can assemble Rubisco in the absence of RbcX likely because Rubisco assembly factor 1 (Raf1) can fulfill its function (Emlyn-Jones et al., 2006). Recently a Raf1 homolog was discovered to be essential for Rubisco assembly in maize (Feiz et al., 2012). This finding suggests that tobacco Raf1 might be able to fulfill the function of its counterpart in cyanobacteria.
CcmM35 is essential for the organization of Rubisco within carboxysomes (Long et al., 2011). Each CcmM35 contains three SSU-like domains which are thought to link adjacent Rubisco holoenzymes and are required to initiate carboxysome assembly (Long et al., 2011; Cameron et al., 2013).
We recently reported two tobacco chloroplast transformed lines, named SeLSX and SeLSM35, where the tobacco Rubisco large subunit gene, rbcL, was replaced with operons containing cyanobacterial Rubisco genes for the large and small subunits along with either rbcX or ccmM35 (Lin et al., 2014a). Although these transformants were able to perform carbon fixation with cyanobacterial Rubisco and grow under elevated CO2, it was not known whether the RbcX and CcmM35 proteins were essential for the assembly and stability of the cyanobacterial Rubisco in chloroplasts. In addition, the poor growth of these transformants even under 0.9% CO2 suggested that future transformants could benefit from improved expression of cyanobacterial Rubisco through better designs of the transgene operons. To address these issues, we generated two additional tobacco chloroplast transformants. In the transformant named SeLS, the two cyanobacterial Rubisco subunits were produced without RbcX or CcmM35, whereas in SeLSYM35 line, we fused YFP to the N-terminus of CcmM35 and optimized the codons of the fused yfp-ccmM35 gene for the chloroplast translation system. We demonstrate that altering the terminator sequences leads to increased accumulation of RNA encoding the cyanobacterial rbcS, which is located 3’ to the rbcL gene in our transgene operons. The improved transgene operons resulted in enhanced Rubisco expression and more rapid growth of the transgenic plants which fixed carbon using the cyanobacterial Rubisco. Thus, neither RbcX or CcmM35 are needed for cyanobacterial Rubisco assembly or vigorous growth under elevated CO2.
RESULTS
Engineering of the tobacco chloroplast genome with synthetic cyanobacterial operons
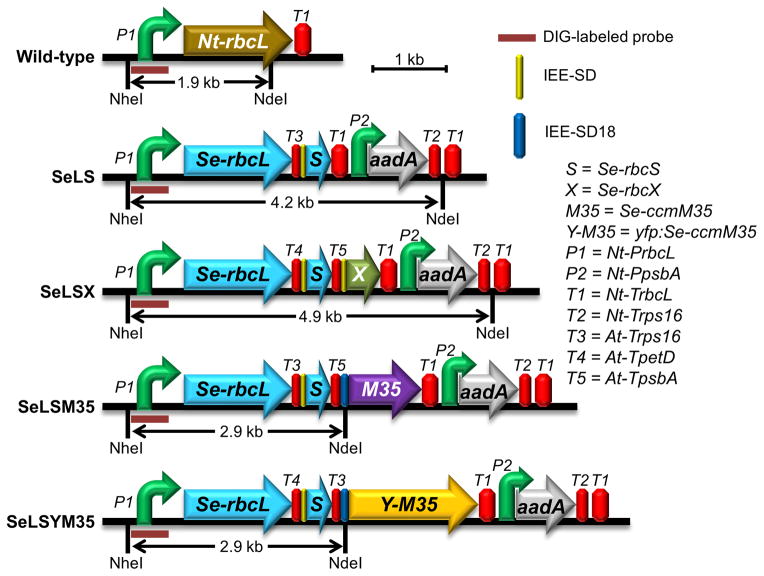
The synthetic operons in the two new transformants possess similar architecture to the previous ones with a terminator, an intercistronic expression element (IEE) and a Shine-Dalgarno sequence (SD) occupying the intergenic regions (Figure 1). Such an arrangement has been shown to result in reliable processing of the transcripts for successful translation of downstream genes inside chloroplasts (Lu et al., 2013; Lin et al., 2014a). Three terminators from the Arabidopsis chloroplast and the native rbcL terminator (Nt-TrbcL) were paired with different genes. The ccmM35 gene in SeLSM35 line and the yfp-ccmM35 gene in SeLSYM35 line are each preceded with the “SD18” translation signal, which has three tandem Shine-Dalgarno sites for improved translation efficiency (Drechsel and Bock, 2011). We confirmed the homoplasmy of the chloroplast genomes in the transformants with a DNA blot (Figure S1a). The complete absence of the native rbcL transcript in the RNA blot also confirmed the successful gene replacement in all four transformants (Figure S1b).
Figure 1.
Comparison of the gene arrangements of the rbcL loci in the wild-type, SeLS (GenBank accession number: KT203393), SeLSX (KM102745), SeLSM35 (KM102746) and SeLSYM35 (KT203394) tobacco lines. Different combinations of terminators from the chloroplast genome of Arabidopsis thaliana (T3–T5), intercistronic expression elements (IEE) and Shine Dalgarno sequences (SD or SD18) are inserted between the cyanobacterial genes. The abbreviations used for the cyanobacterial genes (S, X, M35 and Y-M35), promoters (P1 and P2) and terminators (T1–T5) are listed on the right side of the figure along with the symbols for DIG-labeled probe, IEE-SD and IEE-SD18 regulatory elements. The restriction sites NheI and NdeI that were used in the DNA blot analysis along with the lengths of the expected DNA fragments detected by the DIG-labeled probe are also displayed.
The use of different regulatory elements in the transformed tobacco lines alters the expression of transgenes
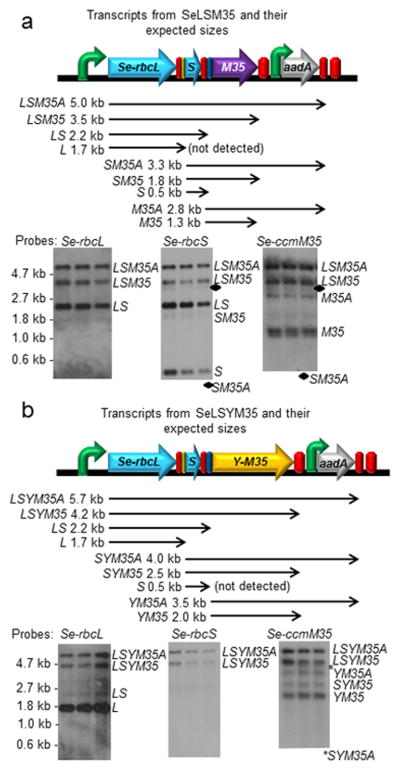
Analyses of the RNA transcripts from the transgene operons show that multigene transcripts are present in all RNA blots, indicating that the IEE sites are only partially processed (Figures 2, 3). Nevertheless, successful production of Rubisco complexes and CcmM35 proteins indicates that downstream genes are still being translated efficiently (Figure 4). We found that the transcripts starting at downstream genes such as Se-rbcS and Se-rbcX were significantly less abundant than those starting at the Se-rbcL gene. The aadA transcript produced from the Nt-PpsbA promoter immediately upstream is highly abundant in all four transgenic lines (Figure S1c).
Figure 2.
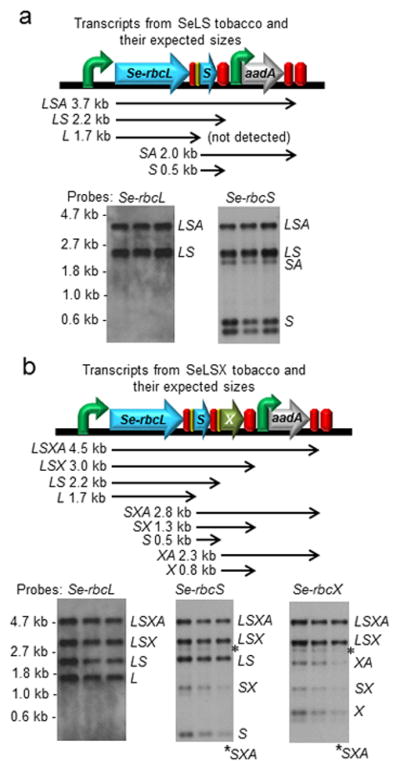
RNA blot analyses of the transcripts derived from the rbcL locus in (a) SeLS and (b) SeLSX tobacco plants. Each blot was run with triplicate samples obtained from three different plants grown under the same conditions. The gene arrangement at the rbcL locus, the expected RNA transcripts and their sizes are displayed on top of the RNA blots for each transformant. Each transcript is named with the abbreviations of the transgenes included in that particular transcript (L = Se-rbcL, S = Se-rbcS, A = aadA, X = Se-rbcX, M35 = Se-ccmM35 and YM35 = YFP:Se-ccmM35). The probes used to detect specific transgenes are indicated on top of each blot. The locations of markers and their sizes are shown on the left side, and the transcripts identified are indicated on the right side of the bands in each blot.
Figure 3.
RNA blot analyses of the transcripts derived from the rbcL locus in (a) SeLSM35 and (b) SeLSYM35 tobacco plants. Each blot was run with triplicate samples obtained from three different plants grown under the same conditions. The gene arrangement at the rbcL locus, the expected RNA transcripts and their sizes are displayed on top of the RNA blots for each transformant. Please refer to Figure 2 for the nomenclature of the transcripts.
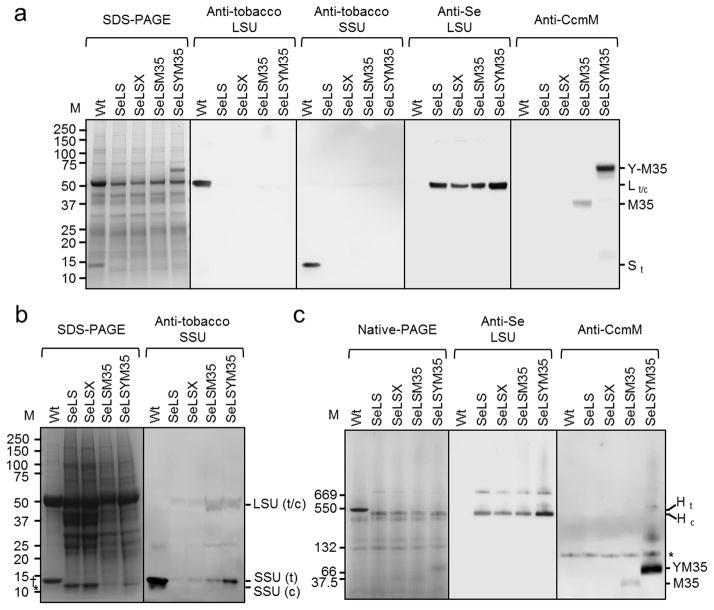
Figure 4.
Expression of Se7942 Rubisco in the four chloroplast transformants as detected by denaturing and non-denaturing polyacrylamide gel electrophoresis. (a) SDS-polyacrylamide gel stained with Coomassie (far left) together with immunoblots probed with an anti-tobacco LSU, anti-tobacco SSU, anti-Se LSU and anti-CcmM antisera. In all cases, 15 μg of total leaf protein from the indicated sources were loaded. (b) Tobacco SSU was detected in the immunoblot of Rubisco samples extracted from wild-type and the four tobacco chloroplast transformant lines. Rubisco complexes were extracted with Triton X-100 and concentrated using HiTrap-Q anion-exchange columns. (c) Native polyacrylamide gel stained with Coomassie Blue (far left) together with immunoblots probed with anti-Se LSU and anti-CcmM antisera. In all cases, 20 μg of total leaf protein from the indicated sources were loaded. Indicated bands correspond to tobacco Rubisco holoenzyme (Ht); Se7942 Rubisco holoenzyme (Hc); YFP-CcmM35 (YM35); CcmM35 (M35) and an unknown cross-reacting protein from tobacco (*). The mass of protein standards (M) are indicated (thyroglobulin (669 kDa); wheat Rubisco (550 kDa); BSA dimer (132 kDa); BSA (66 kDa); CcmM35His (37.5 kDa).
One function of terminators is to stabilize the transcript upstream. Out of the three terminators from Arabidopsis used in this study, we could not detect any transcript ending with the At-Trps16 terminator, which is used in three of our transgene lines, SeLS, SeLSM35 and SeLSYM35. This observation indicates that the At-Trps16 terminator sequence used in our study does not perform well in stabilizing the upstream transcripts.
SDS-PAGE revealed bands of expected masses for the cyanobacterial and tobacco Rubisco large subunits (LSUs) and small subunits (SSUs) in agreement with published data (Chapman et al., 1986; Long et al., 2007) (Figure 4). The presence of the Rubisco subunits and the two forms of CcmM35 proteins (with or without YFP) was confirmed by immunoblotting SDS-polyacrylamide gels with polyclonal antibodies raised against tobacco LSU, cyanobacterial LSU, tobacco SSU and CcmM. Expression of YFP-CcmM35 is higher in SeLSYM35 compared to the level of CcmM35 in the SeLSM35 line (Figure 4c), perhaps due to the codon optimization of the yfp:ccmM35 coding region. Coincidentally, the SeLSYM35 tobacco line also produced the highest amount of cyanobacterial LSU (Figure 4a), probably due to the high abundance of the corresponding transcript as well as the stabilizing effect of YFP-CcmM35 in that line.
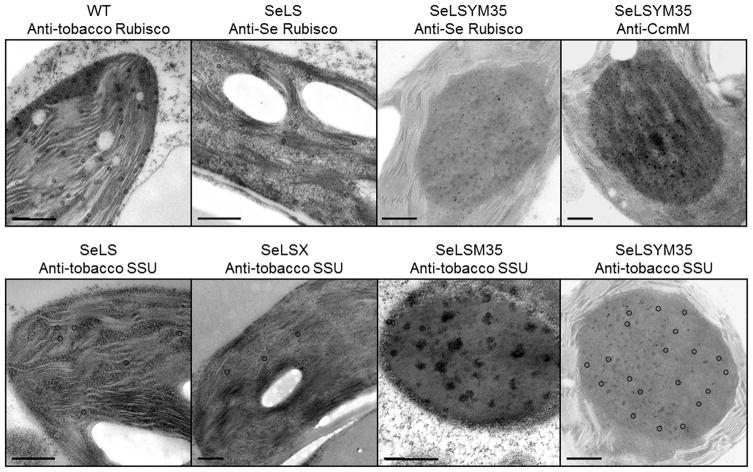
Consistent with previous work, we could not detect tobacco SSU in the total leaf proteins from all four transgenic tobacco lines (Figure 4a), likely due to its instability in the absence of a compatible LSU (Kanevski et al., 1999; Whitney and Andrews, 2001; Lin et al., 2014a). However, by partial purification of Rubisco following extraction with Triton X-100 and concentration by anion-exchange chromatography, we were able to detect a small amount of tobacco SSU in the Rubisco samples from the transformants, particularly in SeLSM35 and SeLSYM35 (Figure 4b), indicating that some hybrid Rubisco enzymes containing both the cyanobacterial LSU and the tobacco SSU may have assembled in these transformants. In electron micrographs of the SeLSM35 and SeLSYM35 lines, the tobacco SSU is seen to be associated with the large complexes formed by the cyanobacterial Rubisco and CcmM35 (Figure 5).
Figure 5.
Localization of Rubisco in the chloroplast stroma of the wild-type and the four transplastomic tobacco lines. Electron micrographs of ultrathin sections of leaf mesophyll cells prepared by high pressure freeze fixation and freeze substitution. Ultrathin sections were probed with the indicated primary antibody and a secondary antibody conjugated with 10 nm gold particles (black circles). Scale bars = 500 nm.
Expression of CcmM35 results in aggregates of Rubisco
Non-denaturing acrylamide gel electrophoresis revealed bands consistent with the predicted molecular weight of the hexadecameric Rubisco holoenzyme from both tobacco (~ 540 kDa) and Se7942 (~ 520 kDa) made up of eight LSUs and eight SSUs (Figure 4c). The composition of the transgenic and wild-type holoenzymes as well as the presence of CcmM35 and YFP-CcmM35 in the SeLSM35 and SeLSYM35 lines were confirmed by immunoblots. Although the formation of such Rubisco holoenzyme is to be expected in SeLS and SeLSX lines, the prevailing models and evidence indicate that CcmM35 may connect a large number of Rubisco complexes into a single and extensive aggregate through interactions between LSUs and SSU-like domains present in CcmM35 (Long et al., 2011). We suspect that the treatment of our samples with Triton X-100 prior to gel electrophoresis partially disrupted such interactions, promoting the formation of the hexadecamer complexes observed on the gel. Even with Triton X-100, we were unable to completely solubilize these complexes formed between CcmM35 and Rubisco.
The formation of large Rubisco aggregates in the presence of either type of CcmM35 was observed by transmission electron microscopy (TEM) with immunogold labelling (Figure 5) (Lin et al., 2014a). In the SeLSYM35 line, the fluorescent image of the leaf tissue also displayed large spherical aggregates consistent with those detected by TEM (Figure S2).
The transformed tobacco plants display photosynthetic performance consistent with the kinetic properties of cyanobacterial Rubisco
The kinetic parameters of the Rubisco extracted from leaves of SeLS and SeLSX (Table 1) were the same as those reported in the literature for the native enzyme extracted from the cyanobacterium Synechococcus PCC7942 ( and ) (Whitehead et al., 2014). Both the maximum catalytic rates ( ) and Michaelis constants ( ) for the enzymes extracted from SeLSM35 and SeLSYM35 were lower, probably due to the effect of minor amounts of tobacco SSU in the enzymes in these lines. The values for the specificity factors are consistent with published values, as were the kinetic properties of tobacco Rubisco, measured contemporaneously, validating our experimental approach (Whitney et al., 1999; Whitney and Andrews, 2001).
TABLE 1.
| Rubisco | (s−1) | (s−1) | (μM) | (μM) | Sc/o | Rubisco μmol sites m−2 |
|---|---|---|---|---|---|---|
| Tobacco | 3.9 ± 0.2 | 1.38 ± 0.04 | 9.0 ± 0.3 | 292 ± 20 | 91.2 ± 6.3 | 13.9 ± 3.5 |
| SeLS | 14.4 ± 0.4 | 1.22 ± 0.32 | 172 ± 64 | 585 ± 38 | 43.0 ± 4.0 | 1.1 ± 0.7 |
| SeLSX | 14.1 ± 1.4 | 1.41 ± 0.49 | 169 ± 39 | 669 ± 132 | 41.1 ± 2.2 | 1.3 ± 0.5 |
| SeLSM35 | 9.9 ± 1.1 | 0.50 ± 0.12 | 139 ± 11 | 429 ± 101 | 60.9 ± 3.0 | 1.3 ± 0.2 |
| SeLSYM35 | 8.9 ± 0.9 | 0.71 ± 0.13 | 140 ± 18 | 534 ± 100 | 48.1 ± 2.2 | 2.1 ± 0.5 |
Table 1. Kinetic properties and abundance of Se7942 Rubisco. Kinetic parameters Se7942 Rubisco extracted from transgenic tobacco lines (SeLS, SeLSX, SeLSM35 and SeLSM35Y) grown in air supplemented with 0.9% (v/v) CO2 and of Rubisco from wild-type tobacco grown at ambient CO2, are shown. and : rate constants for carboxylase and oxygenase activities respectively; and : Michaelis constants for CO2 and O2 respectively; Sc/o: specificity factor. The Rubisco active site concentrations (μmol sites m−2) apply to the leaves used for measurement of photosynthetic CO2 assimilation, shown in Figure 6. The kinetic parameters and active site concentrations are the mean ± sd of three independent determinations, and the Sc/o is the mean ± sd of five replicate determinations.
We also determined the CO2 dependence of photosynthesis (A-Ci) for all tobacco lines, both in normal air (Figure 6) and with a 10-fold reduction in ambient oxygen that would suppress photorespiration, together with the corresponding CO2 compensation points (Figure S3). Expressed on a leaf area basis, it is clear that wild type tobacco had higher rates of net photosynthesis than any of the transgenic lines, at all intercellular CO2 concentrations (Ci) (Figure 6). Suppression of photorespiration under diminished atmospheric O2 was greater in the control than in the transgenic lines, as evident from the accompanying stimulation of photosynthesis at non-saturating levels of CO2 (Figure S3). When the rate of CO2 assimilation was expressed relative to the corresponding concentration of Rubisco catalytic sites (Figure 6), the contrasting properties of the tobacco and cyanobacterial forms of Rubisco were evident: the tobacco enzyme saturating at 500 μmol. intercellular CO2 mol air−1 with a rate of less than 2 s−1, while turnover by the cyanobacterial counterpart continued to increase linearly across the entire range of CO2 exceeding the rate of tobacco, although remaining well below the theoretical maximum of 14 s−1 in the absence of O2 [(Whitehead et al., 2014) and Table 1] even at the highest level of CO2. These observations fully support the respective rates and substrate affinity parameters in Table 1, since substitution of these parameters into the biochemical model of leaf photosynthesis of Farquhar et al. (Farquhar et al., 1980) generated curves which approximated the experimental data (Figure 6).
Figure 6.
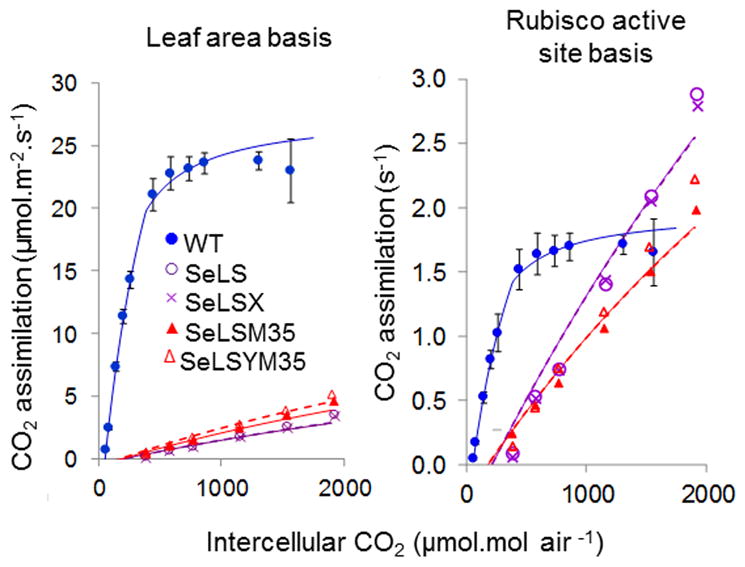
Rates of CO2 assimilation using attached leaves of wild-type and transgenic tobacco lines. Results are expressed relative to leaf area (left); and Rubisco active site concentration (right). The illustrated (solid) lines were generated using a biochemical model (Farquhar et al., 1980) incorporating the kinetic parameters of Table 1 determined for wild-type (WT, blue), SeLS (purple) and SeLSYM35 (red) tobacco lines. The modelled curves were also optimized to minimise deviation from the observed points by varying the leaf Rubisco content (broken lines). Data points are the mean values from three plants per line. Error bars indicate standard error, (omitted for clarity from the transformed lines, but shown in Figure S3).
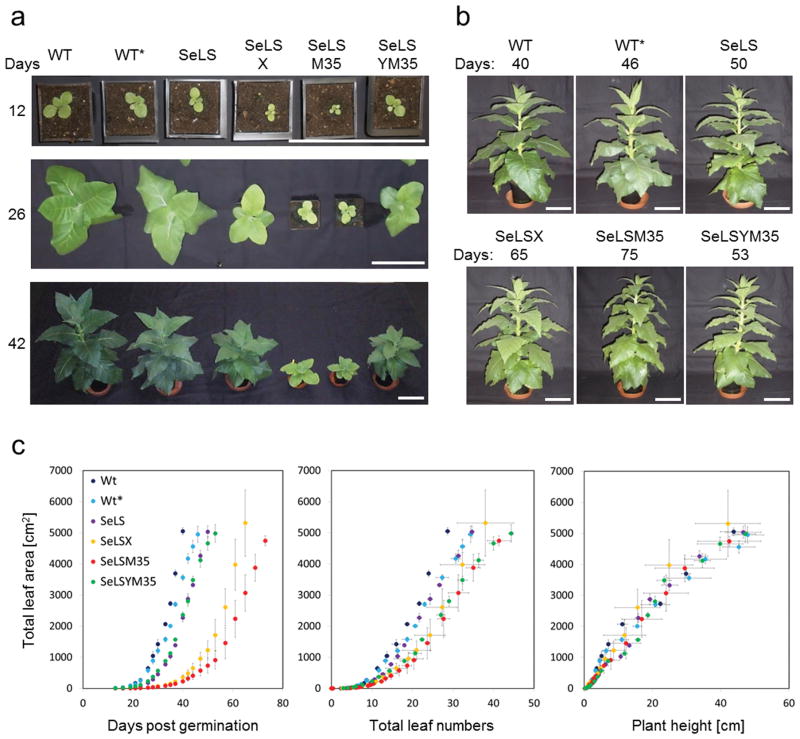
The new transformants grow substantially faster than the previously reported transformants even in the absence of cyanobacterial assembly factors
As expected from the high CO2 compensation points for the transplastomic tobacco plants (Fig S3) these plants could barely survive, let alone grow, at ambient levels of atmospheric CO2. The growth and morphological characteristics of lines expressing Se7942 Rubisco were therefore investigated during growth in air supplemented with 3% (v/v) CO2. The two new transgenic lines exhibited substantially improved growth compared to the original transgenic lines, with the growth rate of SeLS approaching that of wild-type in 3% CO2 despite lacking both RbcX and CcmM35 (Figure 7). The SeLS and SeLSYM35 lines reached the same chosen end-point (immediately preceding anthesis, when the lines had a total leaf area of ~5,000 cm2 per plant) as the controls grown at 3% CO2 only 4 to 7 days later, whereas the SeLSX and SeLSM35 lines reached the same developmental stage 19 and 27 days later, respectively (Figure 7b). Acclimation of the wild-type plants to 3% CO2 (Miller et al., 1997; Schaz et al., 2014) delayed them by about 6 days, relative to plants grown at ambient CO2 (Figure 7b). Furthermore, the wild-type tobacco plants grown at ambient CO2 (400 μmol. CO2 mol air−1) were slightly shorter and showed a lower number of leaves at equivalent values of leaf area (Figure 7c). The leaf distribution of SeLS tobacco plants was indistinguishable from that of the wild-type plants, whereas the other three transformants had numerous smaller leaves at equivalent values of total leaf area (Figure 7c). However, all transformants expressing cyanobacterial Rubisco displayed total leaf areas similar to those of the wild-type plants at comparable heights (Figure 7c).
Figure 7.
Appearance and growth characteristics of wild-type and transgenic tobacco lines expressing Se-Rubisco (a) Wild-type tobacco was grown at normal (400 ppm) and 3% (v/v) CO2 (denoted by “*”) while the SeLS, SeLSX, SeLSM35 and SeLSYM35 lines were all grown in air containing 3% (v/v) CO2. Pictures show the plants at the indicated times (days) after germination. (b) Each plant was pictured at the indicated times, when the total leaf area reached ~5,000 cm2. Scale bars (white): 15 cm throughout. (c) From left to right: the increase in leaf area with time; the increase in leaf area with leaf number; and the increase in leaf area with plant height. Three weeks after germination, the total leaf area, number of leaves and plant height were recorded every 2–3 days until a total plant leaf area of ~5,000 cm2 was attained. The data are expressed as mean values ± sd of three plants per line.
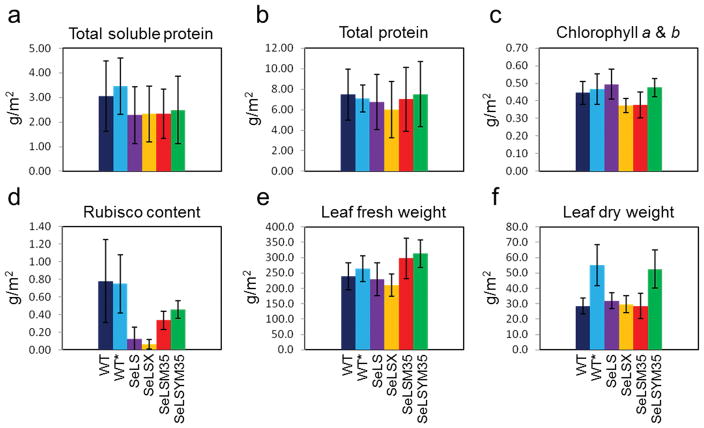
The SeLS transformant displayed dramatically higher efficiency in Rubisco investment
We determined leaf total protein, soluble protein, chlorophyll, Rubisco, fresh and dry mass, from plants at a similar developmental stage (total leaf area ~5,000 cm2 plant−1, pre-anthesis) (Figure 8). These constituents were also measured in leaves at three different positions on the tobacco shoots (youngest fully expanded (top), oldest non senescent (bottom) and intermediate leaves (middle) (Figures S4 and S5). In general, the amounts of soluble protein in the four transgenic lines were lower than those in wild-type tobacco controls. The amounts of total protein in the two lines expressing CcmM35 (SeLSM35 and SeLSYM35) were similar to those in the controls, whereas they were lower in SeLS and SeLSX. The total chlorophyll contents were higher in SeLS and SeLSYM35, which also grew faster than the other transformants. More importantly, all four tobacco transformants, particularly SeLS and SeLSX, produced substantially less cyanobacterial Rubisco than the wild-type Rubisco in the control plants (Figure 8d). Remarkably, the SeLS plants with up to 10-fold less Rubisco (Table 1, Figure 8d) were able to achieve growth rates approaching those of the wild-type plants, indicating the potential benefits of utilizing an inherently faster Rubisco.
Figure 8.
Compositional analyses of transgenic tobacco lines and wild-type controls. Wild-type tobacco (Wt) grown at atmospheric CO2 (400 ppm) together with wild-type tobacco (Wt*) and four transgenic lines (SeLS; SeLSX; SeLSM35; and SeLSYM35) grown in air containing 3% (v/v) CO2 are indicated. (a) total soluble protein; (b) total protein; (c) chlorophyll content; (d) Rubisco content; (e) leaf fresh weight; (f) dry weight. The data are means ± sd of 9 leaves from different positions in the plant canopy ranging from the youngest fully expanded to the oldest non-senescent leaf, collected from each of three plants per genotype.
As expected, the amount of protein (including Rubisco) declined from the youngest fully expanded to the oldest non-senescent leaves (Figures S4 and S5). SeLSYM35 and SeLSM35 had higher Rubisco contents than SeLS and SeLSX, and the difference became more pronounced in the intermediate and oldest non-senescent leaves (Figure S5a). This suggests that association with CcmM35 can inhibit the degradation of cyanobacterial Rubisco. The fresh weights per unit leaf area in SeLSM35 and SeLSYM35 were higher than even the control plants (Figure S5b). Relative to SeLSM35, the faster-growing SeLSYM35 exhibited greater dry weight, which was close to the value measured for the wild-type tobacco grown at the same CO2 concentration (Figure S5c).
DISCUSSION
Successful assembly of Rubisco in chloroplasts involves complex machinery for multiple molecular processes ranging from folding and assembly to functional maintenance (Hauser et al., 2015). β-carboxysomes of cyanobacteria contain Form-IB Rubisco as do higher plant chloroplasts, and so cyanobacterial Rubisco has a good chance of functioning correctly in higher plant chloroplasts with minimal engineering (Cheng et al., 2008; Whitehead et al., 2014). However, it was not known whether the Rubisco from cyanobacteria would be compatible with plant chloroplast assembly systems. Based on the tobacco chloroplast transformants reported previously, it was not clear whether the cyanobacterial assembly factor RbcX or the β-carboxysomal protein CcmM35 was absolutely necessary in chloroplasts (Lin et al., 2014a). In the current work, an additional tobacco transformant, SeLS, expressing only the two cyanobacterial Rubisco subunits without RbcX and CcmM35 was included. Rubisco extracted from the SeLS tobacco plants was found to have the predicted molecular weight for hexadecameric holoenzyme (Figure 4c) and kinetic parameters consistent with cyanobacterial Rubisco (Table 1). These results clearly demonstrate that Se7942 Rubisco can be properly assembled by the tobacco chloroplast chaperones without the intervention of either cyanobacterial RbcX or CcmM35.
Modification of regulatory elements within the synthetic transgene operon lead to slightly enhanced Rubisco expression in SeLS plants compared to SeLSX plants when grown in 3% CO2. Although we do not know the exact reason for higher Rubisco levels in SeLS plants, we suspect that the simpler operon design or the more robust terminator, Nt-TrbcL, at the end of the Se-rbcS gene resulted in the increase in Se-rbcS transcripts (transcript "S" in Figure 2) that we observed. SeLS plants grow faster than other transformants and only slightly more slowly than the wild-type plants under a 3% CO2 atmosphere (Figure 7).
CcmM35 appears to impede the degradation of cyanobacterial Rubisco by chloroplast proteases as the leaves age (Figure S5a). Despite greater cyanobacterial Rubisco abundance, SeLSM35 and SeLSYM35 plants do not grow more rapidly than SeLS. Although association with CcmM35 or tobacco SSU seems to have a slightly negative effect on cyanobacterial Rubisco kinetics, it is unlikely that this effect alone is responsible for the poorer growth of SeLSM35 and SeLSYM35 plants. Slower remobilization of Rubisco-CcmM35 complexes from aging leaves may also impair growth and development. We also suspect that organization of the cyanobacterial enzyme by CcmM35 into extensive complexes larger than 2 μm in size limits access of the substrates to active sites located in the complex interior, leading to reduced rates of photosynthesis and a commensurate underestimation of Rubisco content as determined by 14C-CABP binding. As a comparison, β-carboxysomes found in Se7942 are normally about 100–200 nm in size (Orus et al., 1995; Cannon et al., 2001). Hence, it is likely to be critical to tightly control the size of “carboxysomes” in future transformed plants. This may be achieved through co-expression of other β-carboxysomal components such as the shell proteins, internal components and carbonic anhydrase in precise ratios.
The chloroplast transformation technology used in the current work has the capacity to introduce multiple transgenes and appears ideal for the expression of β-carboxysomes or other CCMs in higher plant chloroplasts to improve photosynthesis (Lu et al., 2013). The discovery of the intercistronic expression element (IEE) has greatly facilitated the stacking of multiple transgenes in synthetic operons for reliable expression of downstream genes in the chloroplast genome (Zhou et al., 2007; Kolotilin et al., 2013; Lu et al., 2013). Although the processing at the IEE sites was incomplete in our transformants, it is not surprising that the genes located downstream were still efficiently translated since proteins from unprocessed multigene transcripts are often successfully produced from tobacco chloroplast transformants (Quesada-Vargas et al., 2005; Whitney et al., 2015). Post-transcriptional control is crucial for chloroplast gene expression; genes located downstream of a large operon can still be efficiently expressed through a strong translational signal (Stern et al., 2010; Hanson et al., 2013).
The unfavourable kinetics of the central carbon-fixing enzyme, Rubisco, necessitates the investment of considerable amounts of nitrogen in this single enzyme. Thus, improving the nitrogen use efficiency of agricultural crops through strategic engineering of Rubisco has long been an important goal in plant biotechnology. Our study clearly shows that even when very modest amounts of cyanobacterial Rubisco are exposed to high CO2 concentrations, the transformed ‘host’ plants are capable of surprisingly fast growth. While this was achieved by means of elevated atmospheric CO2, introduction of a functional cyanobacterial CCM should provide the needed CO2 concentration around the cyanobacterial Rubisco to enable higher rates at normal concentrations of CO2. The current work establishes another critical step towards engineering fully functional carboxysomes into plant chloroplasts.
EXPERIMENTAL PROCEDURES
Construction of the transformation vectors
The amplifications of DNA molecules were carried out with Phusion High-Fidelity DNA polymerase (Thermo Scientific, Grand Island, New York). Table S1 contains the primers ordered from Integrated DNA Technologies (Coralville, Iowa) and used in this work. At-Trps16-IEE-SD-rbcS created in our previous work (Lin et al., 2014a) with overlap extension PCR was digested with MluI restriction enzyme (Thermo Scientific) and ligated in forward orientation at the MauBI site of pCT-rbcL vector (Lin et al., 2014a) to obtain the chloroplast transformation vector pCT-LS used in the generation of SeLS tobacco line.
We designed the Se-ccmM35 and yfp genes with codons optimized for the chloroplast translation system (Puigbo et al., 2007) and had them synthesized by Bioneer Inc. (Alameda, CA). We then used the primers YFPfor-YFPrev and M35CMfor-M35CMrev to amplify the yfp and Se-ccmM35 genes respectively to add 3’ end of IEE-SD sequence in front of yfp and overlaps to join yfp at the N-terminus of Se-ccmM35. We then applied the overlap extension PCR procedure to generate the At-Trps16-IEE-SD18-yfp-ccmM35 DNA fragment, which was then digested with MluI and subsequently ligated into the MauBI site of pCT-rbcL vector to create pCT-rbcL-YM35 vector. At-TpetD-IEE-SD-rbcS was digested with MluI and ligated into the MauBI site of the pCT-rbcL-YM35 vector to obtain the chloroplast transformation vector pCT-LSYM35, used in the generation of SeLSYM35 tobacco line. The procedures to generate tobacco chloroplast transformants and RNA blot analyses are included in the supplemental information.
Generation of transplastomic tobacco plants
We used the Biolistic PDS-1000/He Particle Delivery System (Bio-Rad Laboratories, Inc.) and a tissue culture selection method (Maliga and Tungsuchat-Huang, 2014). Two-week-old tobacco (Nicotiana tabacum cv. Samsun) seedlings germinated in sterile MS agar medium were bombarded with 0.6 μm gold particles carrying the appropriate chloroplast transformation vector. Two days later, the leaves were cut in half and put on RMOP agar plates containing 500 mg/l of spectinomycin and incubated for 4–6 weeks at 23 °C with 14 hours of light per day. The shoots arising from this medium were cut into small pieces of about 5 mm2 and subjected to the second round of regeneration in the same RMOP medium for about 4–6 weeks. If necessary, the shoots from the second round were subjected to another round of selection before they were transferred to MS agar medium containing 500 mg/l of spectinomycin for rooting and then to soil for growth in a greenhouse chamber with elevated atmospheric CO2. DNA blot analyses with the DIG-labeled probe amplified from Nt-PrbcL region were used to determine the homoplasmy of the transformed plants as described previously (Lin et al., 2014a).
RNA blot analyses of the transcripts from transplastomic tobacco plants
First, we generated DNA templates with T7 promoter located on the complement strand at the end of each gene using the primers listed in Table S1. From these DNA templates, the DIG-labeled RNA probes were synthesized with MEGAshortscript kit (Ambion, Foster City, CA) and DIG RNA Labeling Mix (Roche Life Science). Each RNA probe was precipitated with ammonium acetate and ethanol and its concentration was determined with Qubit® RNA BR Assay Kit (Invitrogen, Carlsbad, CA). Generally, as little as 0.1 pg of the probe on a positively charged Nylon membrane can be detected with the alkaline phosphatase-conjugated anti-Digoxigenin and CDP-star chemiluminescent substrate (Roche Life Science).
Tissue samples were collected from fully expanded leaves from the top parts of the plants, rapidly frozen in liquid N2 and stored at −80 ºC before use. These samples were then thawed in RNAlater®-ICE Frozen Tissue Transition Solution (Life Technologies) at −20 ºC. Approximately 30–60 mg of each sample was homogenized in 600 μL of Lysis Buffer from PureLink® RNA Mini Kit (Life Technologies) containing 1% (v/v) 2-mercaptoethanol, and RNA extraction was carried out according to the manufacturer’s protocol. The RNA concentrations were estimated with the Qubit® RNA BR Assay Kit.
For each RNA blot, 0.2 μg of each RNA sample was mixed with three volumes of NorthernMax® Formaldehyde Load Dye (Life Technologies) with 50 μg/mL of ethidium bromide, incubated at 65 ºC for 15 min and separated in a 1.3% agarose denaturing gel prepared with 2% formaldehyde with MOPS buffer under an electric field strength of 7 V/cm for 2 hr. The integrity of the RNA samples in the agarose gel was examined under UV light. The gel was then equilibrated with DEPC-treated H2O for 10 min three times, 50 mM NaOH for 20 min and 20 x SSC buffer for 45 min before the RNAs were transferred to a positively charged Nylon membrane in 20 x SSC under capillary action for 3–5 hr. The RNAs were then crosslinked to the membrane with UV radiation and hybridized with 100 ng of DIG-labeled RNA probe in 3.5 mL of DIG Easy Hyb buffer (Roche Life Science) at 68 ºC overnight. The hybridized probe was then detected with the alkaline phosphatase-conjugated anti-Digoxigenin and CDP-star chemiluminescent substrate (Roche Life Science) according to the manufacturer’s instructions.
Anatomical and biochemical characterization
Transplastomic lines and wild-type tobacco were grown in air containing 3% (v/v) (30,000 ppm) CO2 at a light intensity of 250 μmol photons m−2 s−1 . Duration, temperature and relative humidity during the diel cycle were 16h, 24°C, 70% and 8h, 22°C, 80% for the light and dark periods, respectively. Wild-type tobacco was also grown at normal atmospheric CO2 (400 ppm) under the environmental conditions given above. The leaf area, leaf number and plant height were recorded every 2–3 days using three plants from each genotype. Leaf homogenates were obtained from leaf discs (Andralojc et al., 2014) taken from the lowest non-senescent (bottom), the youngest fully-expanded (top), and mid-way between these extremes (mid) from pre-anthesis plants whose total leaf area was ~5,000 cm2. The total protein (Upreti et al., 2012) and chlorophyll content (Wintermans and de Mots, 1965) were determined using crude leaf homogenates (i.e. prior to centrifugation). The crude homogenates were also used to quantify Rubisco, since significant Rubisco activity was present in insoluble material from leaves expressing SeLSM35 and SeLSYM35. The total Rubisco content in the crude homogenates was investigated by SDS-page comparing the intensity of protein bands (obtained using ImageJ software, NIH USA) with purified Rubisco standards. Soluble protein was determined (Bradford, 1976) following homogenate centrifugation (14,250 x g for 5 min at 4 °C). The leaf fresh weight and leaf dry weight (80°C for 48 hours) were determined using leaf discs from the leaves described above.
SDS-PAGE, blue-native PAGE and immunoblot
For SDS-PAGE and blue-native PAGE, crude leaf homogenates were separated in 4–20% polyacrylamide gradient gels (Thermo Scientific, Horsham, UK) and 3–12% polyacrylamide gradient gels (Invitrogen) respectively, as described previously (Nijtmans et al., 2002; Lin et al., 2014a). The proteins were transfer to a PVDF membrane (Immobilon-P, Millipore, Nottingham, UK) and probed with antibodies against cyanobacterial (SePCC6301) Rubisco, tobacco Rubisco, tobacco Rubisco SSU and CcmM from SePCC7942 (produced by Cambridge Research Biochemicals) as described previously (Lin et al., 2014a). The primary antibodies were detected using an anti-rabbit peroxidase (HRP)-conjugate and a chemiluminescent ECL substrate (Li-Cor, Cambridge, UK).
Cryo-preparation of leaf material, immunogold labelling and transmission electron microscopy
Leaf discs were cryo-fixed using a high pressure freezer EM HPM100 (Leica Microsystems) at a cooling rate of 20,000 Kelvins/sec. The cryo-fixed samples were then subjected to freeze substitution in 0.5% uranyl acetate in dry acetone using an EM AFS unit (Leica Microsystems) and polymerized in Lowicryl HM20 resin (Polysciences) as described previously (Lin et al., 2014b).
Ultrathin sections (60 – 90 nm) of embedded leaf material were subjected to immunogold labelling as describe previously (Lin et al., 2014b). Four primary antibodies against CcmM35, cyanobacterial Rubisco (from Synechococcus elongatus PCC6301), tobacco Rubisco and tobacco Rubisco SSU were used. The primary antibodies were detected using a secondary (goat anti-rabbit) antibody conjugated with 10 nm gold particles (Abcam UK, ab39601). Micrographs were taken using a Jeol 2011 F transmission electron microscope operating at 200kV, equipped with a Gatan Ultrascan CCD camera and a Gatan Dual Vision CCD camera.
Rubisco purification
For and determination, leaf tissue was homogenized in assay buffer (100 mM Bicine-NaOH pH 8.0; 10 mM MgCl2; 1 mM EDTA; 1 mM ε-aminocaproic acid; 1 mM benzamidine; plant protease inhibitor cocktail (diluted 1:100, Sigma); 1mM PMSF; 1 mM KH2PO4; 2 % (w/v) PEG-4000; 10 mM NaHCO3; 10 mM DTT; insoluble PVP (150 mg/gFW)) and was used immediately after centrifugation (2 min, 290 x g). This approach gave results very similar to those obtained when a higher speed centrifugation and G-25 Sephadex desalting was included, prior to assay (Andralojc et al., 2014). The tendency of Rubisco co-expressed with ccmM35 or ccmYM35 to sediment necessitated this simplified approach. Parallel controls demonstrated that the resulting carboxylase activities were entirely RuBP-dependent and were fully inhibited by prior treatment with the Rubisco inhibitor, 2-carboxy-D-arabinitol-1,5-bisphosphate (CABP).
For specificity factor determination, leaf material was homogenized in extraction buffer (40 mM TEA (pH 8, HCl), 10 mM MgCl2, 0.5 mM EDTA, 1 mM K2HPO4, 1 mM ε-aminocaproic, 1 mM benzamidine, 50 mM 2-mercaptoethanol, 5 mM DTT, 10 mM NaHCO3, 1 mM PMSF, 1% (v/v) TX-100 and 1% (w/v) insoluble PVP) and purified using DEAE Sephacel (Pharmacia), a subsequent cycle of anion-exchange chromatography with gradient elution (HiTrap Q, GE-Healthcare), and concentration to ~ 20 mg Rubisco. mL−1 using Ultra-15 centrifugal filter devices (AMICON).
Rubisco activity assay
The determination of and was performed at 25°C in solutions equilibrated with oxygen-free nitrogen or air (79% N2, 21% O2) containing 200 mM Bicine-NaOH pH 8.1, 20 mM MgCl2, 0.4 mM RuBP, 70 units. mL−1 carbonic anhydrase and six different concentrations of sodium bicarbonate ( 3.7 x 1010 Bq mol−1) to provide CO2 concentrations of 7 - 550 μM and 1.4 – 110 μM for cyanobacterial and tobacco Rubisco respectively.
The was obtained from the relationship: . The was obtained from the equation, . The concentration of Rubisco catalytic sites was determined by the [14C]CABP binding method (Yokota and Canvin, 1985) using [21-14C]CABP (3.7 x 1010 Bq mol−1 and 3.7 x 1011 Bq mol−1 for WT and transplastomic tobacco samples, respectively). The specificity factor (Sc/o) was determined at 25°C by monitoring the total consumption of RuBP in an oxygen electrode, as described previously (Parry et al., 1989).
Gas-exchange measurements
The gas exchange measurements were performed using a LI-6400XT portable photosynthesis system (LiCor, Lincoln, Nebraska, USA) at constant irradiance (1,000 μmol photons. m−2. s−1), 25 ± 0.5°C, a vapour pressure deficit of 0.8 – 1.0 kPa, a flow rate of 200 μmol s−1 with CO2 concentrations ranging between 100 and 2,000 μmol. mol air −1. The A-Ci curves were determined under photorespiratory and non-photorespiratory conditions, using air containing 21% and 2% (v/v) O2 respectively. The results were related to both leaf area and Rubisco active site concentration, the latter determined by [14C]-CABP binding assay (Yokota and Canvin, 1985).
Accession Numbers
The sequences of chloroplast transformation constructs used in this study can be found in the GenBank data library under the following accession numbers: SeLS, GenBank accession KT203393; SeLSX, GenBank accession KM102745; SeLSM35, GenBank accession KM102746; and SeLSYM35, GenBank accession KT203394.
Supplementary Material
Replacement of the Nt-rbcl gene with synthetic operons containing cyanobacterial genes.
YFP-CcmM35 bodies in chloroplasts within leaf tissue of the SeLSYM35 tobacco line.
A-Ci curves and CO2 compensation points from attached leaves containing the wild type and cyanobacterial Rubisco expressed in tobacco chloroplasts.
Quantification of total leaf proteins and chlorophylls.
Quantification of Rubisco and leaf fresh and dry weights.
The oligonucleotides used in the construction of chloroplast transformation vectors, DNA blot analyses and RNA blot analyses of the transplastomic tobacco lines.
Acknowledgments
This material is based upon work supported by Biotechnology and Biological Sciences Research Council under grant number BB/I024488/1 to M.A.J.P., the National Science Foundation under grant number EF-1105584 to M.R.H. and the National Institute of General Medical Sciences of the National Institutes of Health under award number F32GM103019 to M.T.L. P.J.A. and M.A.J.P. also acknowledge support from the 20:20 Wheat Institute Strategic Program (BBSRC BB/J/00426X/1). M.R.H. and M.T.L. also acknowledge support from the Cornell University Biotechnology Resource Center (NIH S10RR025502) for the shared Zeiss LSM 710 Confocal. We also acknowledge WenFang Li for the technical assistance in generating the tobacco chloroplast transformants and Cheryl Kerfeld for supplying us with cyanobacterial genomic DNA.
Contributor Information
Alessandro Occhialini, Email: alessandro.occhialini@rothamsted.ac.uk.
Myat T. Lin, Email: mtl84@cornell.edu.
P. John Andralojc, Email: john.andralojc@rothamsted.ac.uk.
Maureen R. Hanson, Email: mrh5@cornell.edu.
References
- Andersson I, Backlund A. Structure and function of Rubisco. Plant Physiology and Biochemistry. 2008;46:275–291. doi: 10.1016/j.plaphy.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Andralojc PJ, Bencze S, Madgwick PJ, Philippe H, Powers SJ, Shield I, Karp A, Parry MAJ. Photosynthesis and growth in diverse willow genotypes. Food and Energy Security. 2014;3:69–85. [Google Scholar]
- Bobik TA. Polyhedral organelles compartmenting bacterial metabolic processes. Appl Microbiol Biotechnol. 2006;70:517–525. doi: 10.1007/s00253-005-0295-0. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical biochemistry. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cameron JC, Wilson SC, Bernstein SL, Kerfeld CA. Biogenesis of a bacterial organelle: the carboxysome assembly pathway. Cell. 2013;155:1131–1140. doi: 10.1016/j.cell.2013.10.044. [DOI] [PubMed] [Google Scholar]
- Cannon GC, Bradburne CE, Aldrich HC, Baker SH, Heinhorst S, Shively JM. Microcompartments in prokaryotes: carboxysomes and related polyhedra. Applied and environmental microbiology. 2001;67:5351–5361. doi: 10.1128/AEM.67.12.5351-5361.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MS, Smith WW, Suh SW, Cascio D, Howard A, Hamlin R, Xuong NH, Eisenberg D. Structural studies of Rubisco from tobacco. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 1986;313:367–378. doi: 10.1098/rstb.1986.0044. [DOI] [PubMed] [Google Scholar]
- Cheng SQ, Liu Y, Crowley CS, Yeates TO, Bobik TA. Bacterial microcompartments: their properties and paradoxes. BioEssays. 2008;30:1084–1095. doi: 10.1002/bies.20830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsel O, Bock R. Selection of Shine-Dalgarno sequences in plastids. Nucleic acids research. 2011;39:1427–1438. doi: 10.1093/nar/gkq978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emlyn-Jones D, Woodger FJ, Price GD, Whitney SM. RbcX can function as a rubisco chaperonin, but is non-essential in Synechococcus PCC7942. Plant & cell physiology. 2006;47:1630–1640. doi: 10.1093/pcp/pcl028. [DOI] [PubMed] [Google Scholar]
- Evans JR. Photosynthesis and nitrogen relationships in leaves of C3 plants. Oecologia. 1989;78:9–19. doi: 10.1007/BF00377192. [DOI] [PubMed] [Google Scholar]
- Farquhar GD, von Caemmerer S, Berry JA. A biochemical model of photosynthetic CO2 assimilation in leaves of C 3 species. Planta. 1980;149:78–90. doi: 10.1007/BF00386231. [DOI] [PubMed] [Google Scholar]
- Feiz L, Williams-Carrier R, Wostrikoff K, Belcher S, Barkan A, Stern DB. Ribulose-1,5-bis-phosphate carboxylase/oxygenase accumulation factor1 is required for holoenzyme assembly in maize. Plant Cell. 2012;24:3435–3446. doi: 10.1105/tpc.112.102012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson MR, Gray BN, Ahner BA. Chloroplast transformation for engineering of photosynthesis. J Exp Bot. 2013;64:731–742. doi: 10.1093/jxb/ers325. [DOI] [PubMed] [Google Scholar]
- Hauser T, Popilka L, Hartl FU, Hayer-Hartl M. Role of auxiliary proteins in Rubisco biogenesis and function. Nature Plants. 2015;1:15065. doi: 10.1038/nplants.2015.65. [DOI] [PubMed] [Google Scholar]
- Kanevski I, Maliga P, Rhoades DF, Gutteridge S. Plastome engineering of ribulose-1,5-bisphosphate carboxylase/oxygenase in tobacco to form a sunflower large subunit and tobacco small subunit hybrid. Plant Physiology. 1999;119:133–142. doi: 10.1104/pp.119.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolotilin I, Kaldis A, Pereira EO, Laberge S, Menassa R. Optimization of transplastomic production of hemicellulases in tobacco: effects of expression cassette configuration and tobacco cultivar used as production platform on recombinant protein yields. Biotechnol Biofuels. 2013;6:65. doi: 10.1186/1754-6834-6-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Occhialini A, Andralojc PJ, Parry MA, Hanson MR. A faster Rubisco with potential to increase photosynthesis in crops. Nature. 2014a;513:547–550. doi: 10.1038/nature13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Occhialini A, Andralojc PJ, Devonshire J, Hines KM, Parry MAJ, Hanson MR. β-Carboxysomal proteins assemble into highly organized structures in Nicotiana chloroplasts. Plant J. 2014b;79:1–12. doi: 10.1111/tpj.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Young AL, Starling-Windhof A, Bracher A, Saschenbrecker S, Rao BV, Rao KV, Berninghausen O, Mielke T, Hartl FU, Beckmann R, Hayer-Hartl M. Coupled chaperone action in folding and assembly of hexadecameric Rubisco. Nature. 2010;463:197–202. doi: 10.1038/nature08651. [DOI] [PubMed] [Google Scholar]
- Long BM, Badger MR, Whitney SM, Price GD. Analysis of carboxysomes from Synechococcus PCC7942 reveals multiple Rubisco complexes with carboxysomal proteins CcmM and CcaA. J Biol Chem. 2007;282:29323–29335. doi: 10.1074/jbc.M703896200. [DOI] [PubMed] [Google Scholar]
- Long BM, Rae BD, Badger MR, Price GD. Over-expression of the b-carboxysomal CcmM protein in Synechococcus PCC7942 reveals a tight co-regulation of carboxysomal carbonic anhydrase (CcaA) and M58 content. Photosynthesis research. 2011;109:33–45. doi: 10.1007/s11120-011-9659-8. [DOI] [PubMed] [Google Scholar]
- Lu Y, Rijzaani H, Karcher D, Ruf S, Bock R. Efficient metabolic pathway engineering in transgenic tobacco and tomato plastids with synthetic multigene operons. Proc Natl Acad Sci USA. 2013;110:E623–632. doi: 10.1073/pnas.1216898110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliga P, Tungsuchat-Huang T. Plastid transformation in Nicotiana tabacum and Nicotiana sylvestris by biolistic DNA delivery to leaves. Methods in molecular biology. 2014;1132:147–163. doi: 10.1007/978-1-62703-995-6_8. [DOI] [PubMed] [Google Scholar]
- McGrath JM, Long SP. Can the cyanobacterial carbon-concentrating mechanism increase photosynthesis in crop species? A theoretical analysis. Plant Physiol. 2014;164:2247–2261. doi: 10.1104/pp.113.232611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A, Tsai CH, Hemphill D, Endres M, Rodermel S, Spalding M. Elevated CO2 effects during leaf ontogeny (A new perspective on acclimation) Plant Physiol. 1997;115:1195–1200. doi: 10.1104/pp.115.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Cajar O, Whitney SM. Evolving improved Synechococcus Rubisco functional expression in Escherichia coli. Biochem J. 2008;414:205–214. doi: 10.1042/BJ20080668. [DOI] [PubMed] [Google Scholar]
- Nijtmans LG, Henderson NS, Holt IJ. Blue native electrophoresis to study mitochondrial and other protein complexes. Methods. 2002;26:327–334. doi: 10.1016/S1046-2023(02)00038-5. [DOI] [PubMed] [Google Scholar]
- Ort DR, Merchant SS, Alric J, Barkan A, Blankenship RE, Bock R, Croce R, Hanson MR, Hibberd JM, Long SP, Moore TA, Moroney J, Niyogi KK, Parry MA, Peralta-Yahya PP, Prince RC, Redding KE, Spalding MH, van Wijk KJ, Vermaas WF, von Caemmerer S, Weber AP, Yeates TO, Yuan JS, Zhu XG. Redesigning photosynthesis to sustainably meet global food and bioenergy demand. Proceedings of the National Academy of Sciences of the United States of America. 2015 doi: 10.1073/pnas.1424031112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orus MI, Rodriguez ML, Martinez F, Marco E. Biogenesis and Ultrastructure of Carboxysomes from Wild Type and Mutants of Synechococcus sp. Strain PCC 7942. Plant Physiol. 1995;107:1159–1166. doi: 10.1104/pp.107.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry MA, Andralojc PJ, Mitchell RA, Madgwick PJ, Keys AJ. Manipulation of Rubisco: the amount, activity, function and regulation. J Exp Bot. 2003;54:1321–1333. doi: 10.1093/jxb/erg141. [DOI] [PubMed] [Google Scholar]
- Parry MAJ, Keys AJ, Gutteridge S. Variation in the specificity factor of C3 higher plant rubiscos determined by the total consumption of ribulose-P2. J Exp Bot. 1989;40:317–320. [Google Scholar]
- Price GD, Pengelly JJ, Forster B, Du J, Whitney SM, von Caemmerer S, Badger MR, Howitt SM, Evans JR. The cyanobacterial CCM as a source of genes for improving photosynthetic CO2 fixation in crop species. Journal of experimental botany. 2013;64:753–768. doi: 10.1093/jxb/ers257. [DOI] [PubMed] [Google Scholar]
- Puigbo P, Guzman E, Romeu A, Garcia-Vallve S. OPTIMIZER: a web server for optimizing the codon usage of DNA sequences. Nucleic Acids Res. 2007;35:W126–131. doi: 10.1093/nar/gkm219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada-Vargas T, Ruiz ON, Daniell H. Characterization of heterologous multigene operons in transgenic chloroplasts: transcription, processing, and translation. Plant Physiology. 2005;138:1746–1762. doi: 10.1104/pp.105.063040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaz U, Dull B, Reinbothe C, Beck E. Influence of root-bed size on the response of tobacco to elevated CO2 as mediated by cytokinins. AoB PLANTS. 2014:6. doi: 10.1093/aobpla/plu010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DB, Goldschmidt-Clermont M, Hanson MR. Chloroplast RNA metabolism. Annual review of plant biology. 2010;61:125–155. doi: 10.1146/annurev-arplant-042809-112242. [DOI] [PubMed] [Google Scholar]
- Tabita FR, Hanson TE, Satagopan S, Witte BH, Kreel NE. Phylogenetic and evolutionary relationships of RubisCO and the RubisCO-like proteins and the functional lessons provided by diverse molecular forms. Philosophical Transactions of the Royal Society of London Series B: Biological Sciences. 2008;363:2629–2640. doi: 10.1098/rstb.2008.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upreti GC, Wang Y, Finn A, Sharrock A, Feisst N, Davy M, Jordan RB. U-2012: An improved Lowry protein assay, insensitive to sample color, offering reagent stability and enhanced sensitivity. Biotechniques. 2012;52:159–166. doi: 10.2144/000113818. [DOI] [PubMed] [Google Scholar]
- Vitlin Gruber A, Nisemblat S, Azem A, Weiss C. The complexity of chloroplast chaperonins. Trends in plant science. 2013;18:688–694. doi: 10.1016/j.tplants.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Whitehead L, Long BM, Price GD, Badger MR. Comparing the in vivo function of α-carboxysomes and β-carboxysomes in two model cyanobacteria. Plant Physiology. 2014;165:398–411. doi: 10.1104/pp.114.237941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, Andrews TJ. Plastome-encoded bacterial ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) supports photosynthesis and growth in tobacco. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:14738–14743. doi: 10.1073/pnas.261417298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, Houtz RL, Alonso H. Advancing our understanding and capacity to engineer nature's CO2-sequestering enzyme, Rubisco. Plant Physiol. 2011;155:27–35. doi: 10.1104/pp.110.164814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, von Caemmerer S, Hudson GS, Andrews TJ. Directed mutation of the Rubisco large subunit of tobacco influences photorespiration and growth. Plant Physiology. 1999;121:579–588. doi: 10.1104/pp.121.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, Birch R, Kelso C, Beck JL, Kapralov MV. Improving recombinant Rubisco biogenesis, plant photosynthesis and growth by coexpressing its ancillary RAF1 chaperone. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:3564–3569. doi: 10.1073/pnas.1420536112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermans JF, de Mots A. Spectrophotometric characteristics of chlorophylls a and b and their pheophytins in ethanol. Biochim. Biophys. Acta. 1965;109:448–453. doi: 10.1016/0926-6585(65)90170-6. [DOI] [PubMed] [Google Scholar]
- Yokota A, Canvin DT. Ribulose bisphosphate carboxylase/oxygenase content determined with [14C]carboxypentitol bisphosphate in plants and algae. Plant Physiol. 1985;77:735–739. doi: 10.1104/pp.77.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarzycki J, Axen SD, Kinney JN, Kerfeld CA. Cyanobacterial-based approaches to improving photosynthesis in plants. Journal of experimental botany. 2013;64:787–798. doi: 10.1093/jxb/ers294. [DOI] [PubMed] [Google Scholar]
- Zhou F, Karcher D, Bock R. Identification of a plastid intercistronic expression element (IEE) facilitating the expression of stable translatable monocistronic mRNAs from operons. Plant J. 2007;52:961–972. doi: 10.1111/j.1365-313X.2007.03261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Replacement of the Nt-rbcl gene with synthetic operons containing cyanobacterial genes.
YFP-CcmM35 bodies in chloroplasts within leaf tissue of the SeLSYM35 tobacco line.
A-Ci curves and CO2 compensation points from attached leaves containing the wild type and cyanobacterial Rubisco expressed in tobacco chloroplasts.
Quantification of total leaf proteins and chlorophylls.
Quantification of Rubisco and leaf fresh and dry weights.
The oligonucleotides used in the construction of chloroplast transformation vectors, DNA blot analyses and RNA blot analyses of the transplastomic tobacco lines.