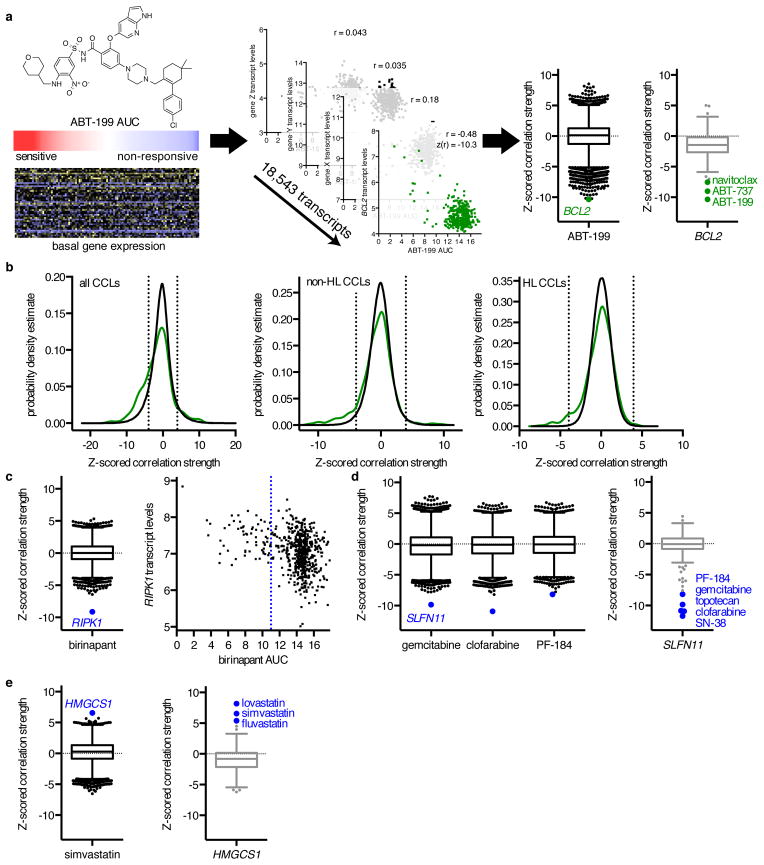
Figure 1. Correlating Gene Expression and CCL Sensitivity Data Illuminates Known Small-Molecule Mechanisms of Action.
(a) Calculation of z-scored Pearson correlation coefficients between small-molecule sensitivity data, expressed as areas under concentration-response curves (AUCs), with basal gene-expression measurements, expressed as log2 robust-multi-array-average values. We examined 18,543 correlation coefficients of transcript levels to ABT-199 sensitivity (black), and 481 correlation coefficients of small molecules to BCL2 expression (gray) across non-hematopoietic and lymphoid (non-HL) CCLs. Box-and-whisker plot outlier points represent Tukey outliers (1.5 × interquartile range). (b) Distribution of z-scored Pearson correlation coefficients between 660 annotated small-molecule–target pairs (green) across all CCLs, non-HL CCLs, and HL CCLs compared to random sampling of correlation coefficients (black). Dashed lines represent two-tailed Bonferroni-corrected significance (|z| = 3.96). (c–e) Expression–sensitivity correlations for target–pathway connections (blue), including (c) the Smac mimetic birinapant; (d) the nucleoside analogues clofarabine and gemcitabine, the IKKβ inhibitor PF-184, and the transcript SLFN11; and (e) the statin simvastatin and the transcript HMGCS1. The blue dashed line represents the low-AUC (left) cutoff for the robust z-score of birinapant AUCs.