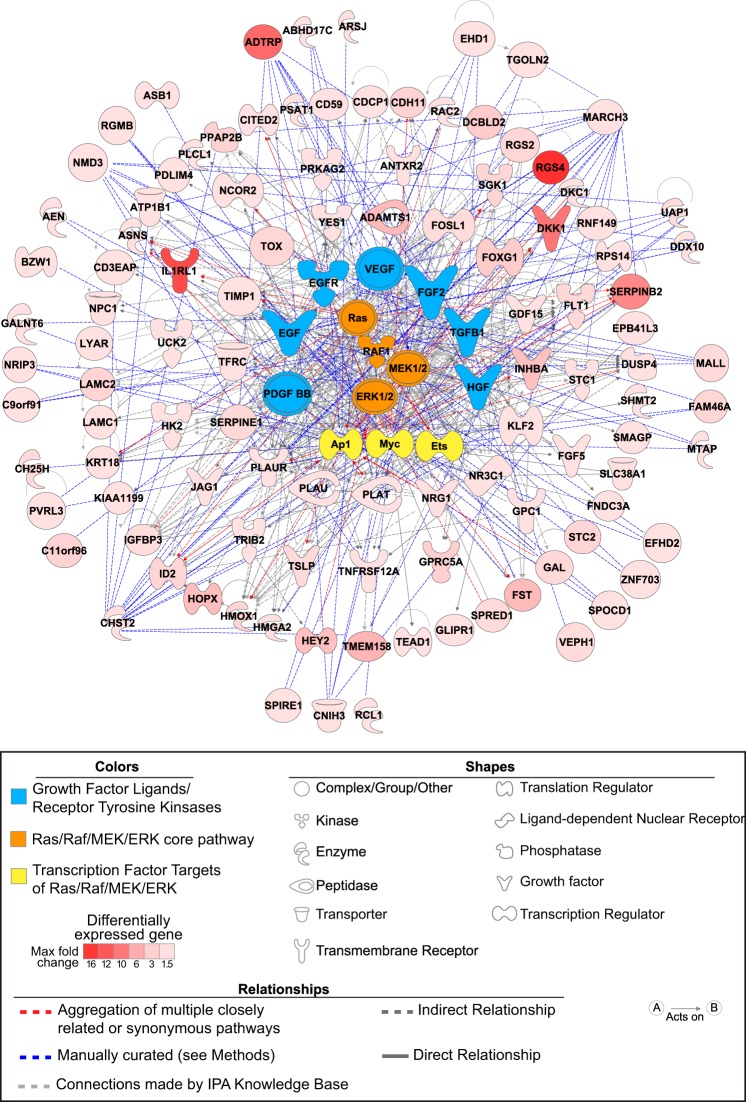
Fig. 4.
Regulatory network derived from the less-than-additive interaction between BMPR2 silencing and mitogenic stimulation. HPAECs expression profiles in the presence or absence of BMPR2 knockdown and/or a mitogen (phorbol 12-myristate 13-acetate, PMA) were examined for significant interactions. Upregulation by both BMPR2 silencing and PMA (FDR < 5%, for both) with less-than-additive combined effects was the predominant pattern identified. Regulatory nodes were chosen based on the following algorithm: 1) IPA-predicted upstream regulators present among the 107 interacting genes; 2) components of the Ras/Raf/MEK/ERK shown to be activated after BMPR2 knockdown and predicated by IPA as upstream regulators (P < 0.001); 3) tyrosine kinase receptors/ligands upstream of Ras/Raf with IPA activation prediction z-scores ≥ 2 and associated with PAH pathogenesis (EGF, EGFR, FGF2, TGFB1, VEGF, HGF, and PDGF BB); and 4) transcription factors downstream of Raf/ERK associated with PAH pathogenesis (AP1, ETS, and MYC family members). The Ingenuity Knowledge Base connected 77 of 107 interacting genes and connections for the remaining 30 genes were manually curated using PubMed, the Molecular Signatures Database (MSigDB) version 4.0, and STRING version 9.1.