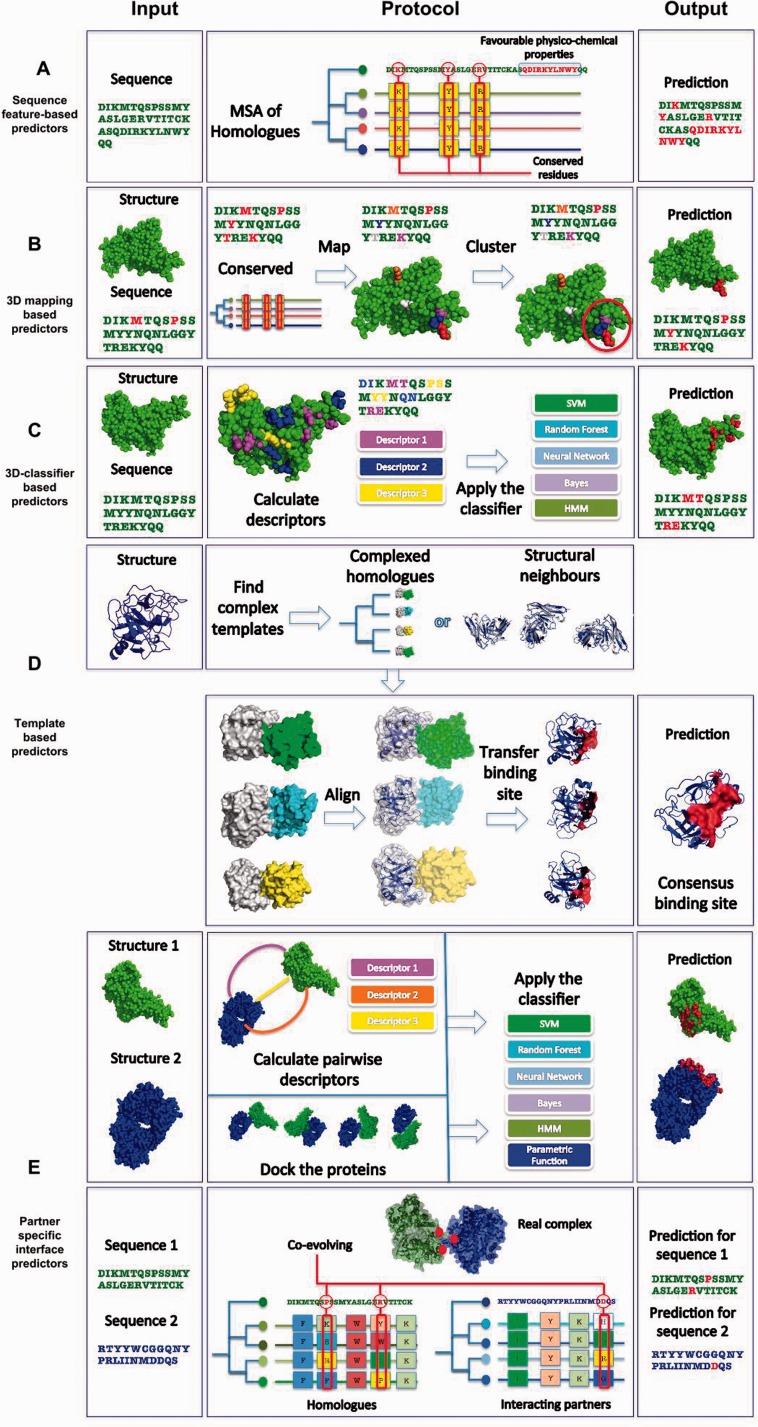
Figure 1.
Classification of existing protein interface prediction methods. In the leftmost column we present the input required by a method. In the middle column, a simplified pipeline for the protocol is presented. In the rightmost, prediction column, the resulting binding site is shown in red. Most methods output a ranked list of possible binding sites. Here for simplicity, we show a single result for each method. (A) Sequence-feature-based predictors: These methods receive a protein sequence. Sequential features of the input are compared with features thought to contribute to a residue being part of an interface, such as conservation scores and physico-chemical properties. (B) 3D mapping-based predictors: These methods receive a protein structure and its sequence as input. Evolutionary conservation is coupled with 3D surface and sequence information. Conserved residues can be grouped according to their surface proximity to form contiguous interface patches. (C) 3D-classifier-based predictors: The input for these methods is a protein structure and its sequence. Distinct sets of attributes (physico-chemical, evolution, 3D structural features, etc.) are used as an input to a learning method such as a SVM or Random Forest. (D) Template-based predictors: These methods receive a protein structure (and thus its sequence) as input. Complex templates are then identified, which can be homologues or structural neighbours (these are shown in white, whereas their binding partners are in green, cyan and yellow). Templates of the input protein are aligned to the query protein. The most commonly aligned contact sites are returned as a prediction. (E) Partner-specific interface predictors: These methods receive the structures/sequences of two proteins that are assumed to interact. The three groups of methods are shown for this category. Partner-specific descriptors can be calculated to predict interfaces. In some cases docking is used to sample possible orientations to identify a consensus binding site. Partner-specific descriptors and docking poses are used as input for parametric functions and classifiers to obtain the final result. In the co-evolution-based strategy, a MSA of interacting homologues is created and sites that appear to mutate in concert (co-evolve) are assumed to constitute the binding site. A colour version of this figure is available at BIB online: http://bib.oxfordjournals.org.