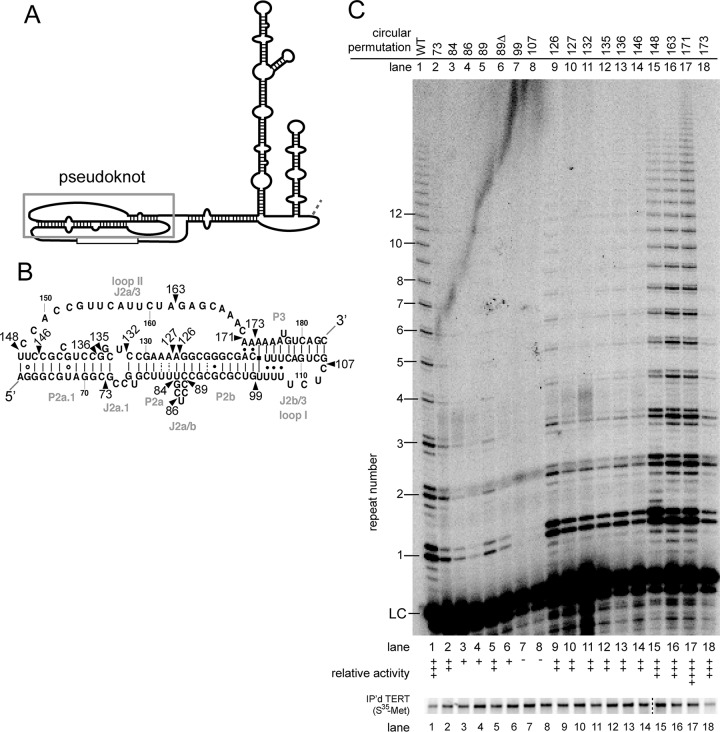
FIG 2.
Circular permutations throughout the pseudoknot cause a wide range of phenotypes. (A) Secondary structure of hTR with the pseudoknot region indicated. (B) Detailed structure of the pseudoknot, based a previously published study (35). Wild-type nucleotide numbers are indicated. Circular permutations are indicated by arrowheads and are named by the corresponding nucleotide at the new 5′ end. (C) In vitro telomerase assays exhibit various levels of activity. LC, internal radiolabeled recovery and loading control. The level of activity was binned as in Fig. 1. Immunopurified [35S]methionine-labeled FLAG-hTERT shown below. Protein samples in lanes 1 to 14 and lanes 15 to 18 were run on two separate gels in parallel, as indicated by the dashed line.