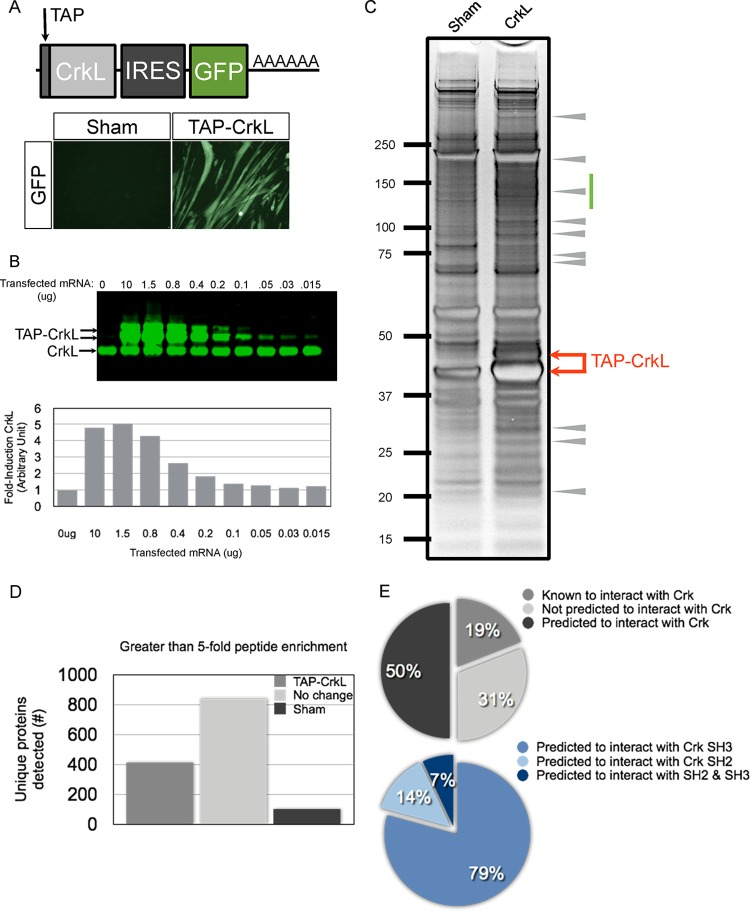
FIG 3.
Identification of CrkL binding proteins from myotube lysates. (A) Cartoon representation of the TAP-tagged CrkL transcript that was tested. The transcript contains an N-terminal TAP tag, the CrkL coding sequence, an IRES-GFP, and an SV40 poly(A) sequence. (B) Myotubes transfected with serial dilutions of TAP-CrkL RNA and cell lysates were analyzed at 18 h posttransfection. CrkL RNA scales linearly over a 5-fold range and permits reconstitution of exogenous CrkL at levels similar to physiological CrkL protein levels. (C) TAP-CrkL proteins were purified from myotube lysates and subjected to SDS-PAGE and silver staining. The entire lane from either the sham or TAP-CrkL condition was cut into 18 individual bands and subjected to MS/MS analysis. Arrows indicate the size of the TAP-CrkL doublet band. The green bar indicates an area of the gel that was enriched with CrkL-specific interactors. Arrowheads indicate clear CrkL-specific proteins. (D) MS/MS summary of proteins identified under the two conditions. There were roughly 1,500 proteins in total, of which ∼400 proteins showed a >5-fold peptide enrichment specific to the TAP-CrkL condition. (E) Pie chart summary of CrkL-interacting proteins, summarizing proteins known, predicted, and not predicted to interact with CrkL. Of the predicted interacting proteins, many are predicted to interact with the SH3 domains of CrkL (79%).