FIG 5.
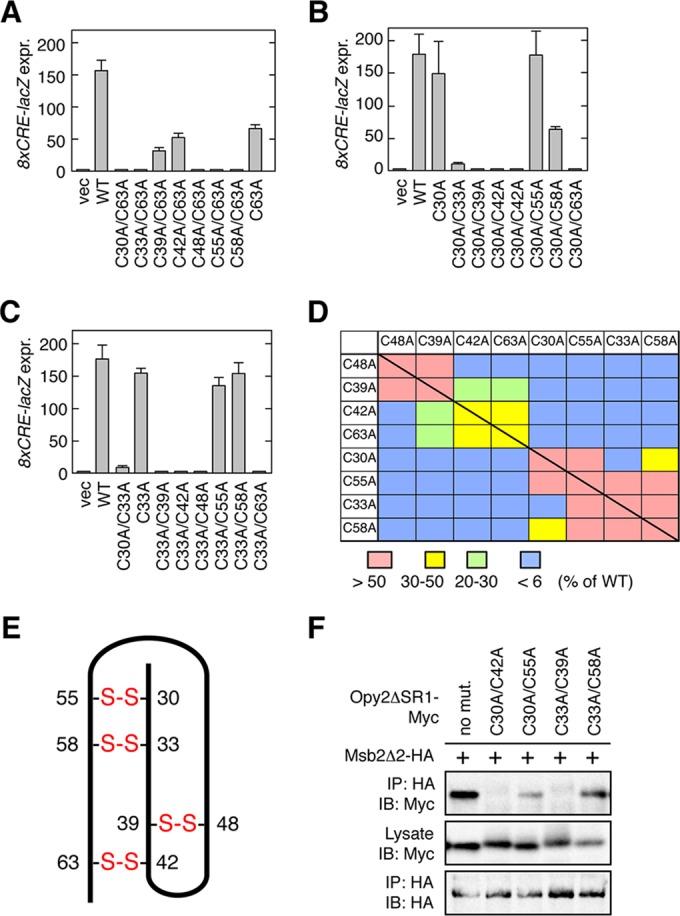
Disulfide bonding pattern of the Opy2 CR domain. (A to C) Osmostress-induced expression of the Hog1-specific reporter gene 8×CRE-lacZ was assayed as described in the legend to Fig. 4D. Error bars represent SDs (n = 3 or 4). (D) Summary of Hog1-specific gene expression assays. Each block (except the blocks on the diagonal line) represents an opy2 double mutant that contains one of the mutations indicated in the top row and one of the mutations indicated in the left column. The blocks on the diagonal line represent single mutants. The strength of reporter gene expression of the mutants was normalized to that of the wild-type control and is indicated in color. (E) Deduced disulfide bonding pattern of the Opy2 CR domain. The bold line represents the peptide chain of the CR domain. The numbers refer to the amino acid positions of the eight cysteine residues. Disulfide bonds are indicated in red. (F) Msb2-Opy2 binding assays. TM257 was cotransformed with expression plasmids for Msb2Δ2-HA (+) and Opy2ΔSR1-Myc (containing the indicated mutations), both of which were under the control of the GAL1 promoter. Cell growth, preparation of cell lysates, immunoprecipitation, and immunoblotting were as described in the legend to Fig. 2A, except for the antibodies used. Buffer A containing 1.0% digitonin was used.
