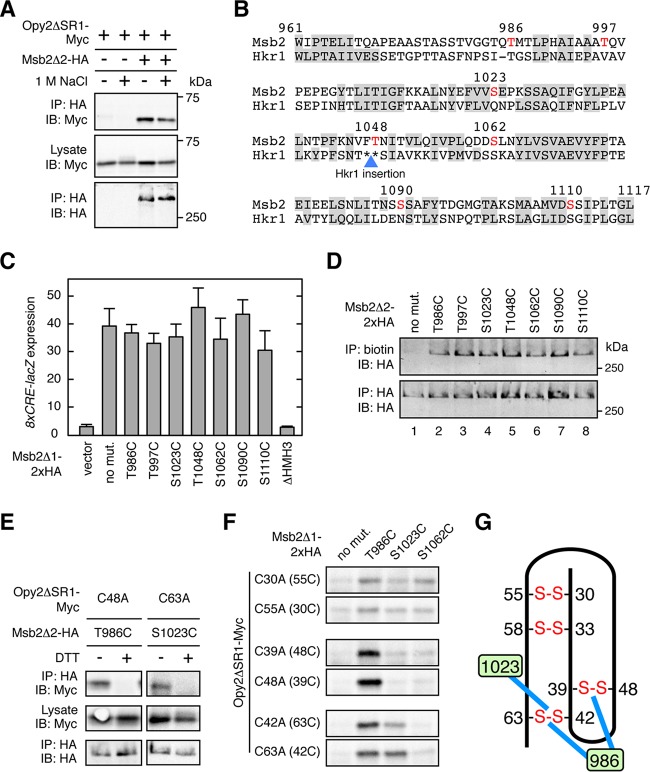
FIG 6.
Direct interaction between the Opy2 CR domain and the Msb2 HMH domain. (A) Effect of osmostress on Msb2-Opy2 binding assays. TM257 was cotransformed with expression plasmids for Opy2ΔSR1-Myc and Msb2Δ2-HA, both of which were under the control of the GAL1 promoter. Cells were grown in CARaf medium, and expression of the tagged Opy2 and Msb2 was induced by 2% galactose for 2 h. The cells were then exposed (+) or not (−) to 1 M NaCl for 5 min. Preparation of cell lysates, immunoprecipitation, and immunoblotting were as described in the legend to Fig. 2A, except for the antibodies used. Buffer A containing 1.0% digitonin was used. (B) Predicted surface accessibility of the amino acid residues in the HMH domain. The HMH domain sequences of Msb2 and Hkr1 are aligned. Residues predicted to be solvent inaccessible are highlighted by gray shading. The blue triangle denotes the site of the 62-amino-acid-residue insertion sequence in Hkr1 relative to the Msb2 sequence. The surface-exposed Ser or Thr residues that were mutated in this work are shown in red. Numbers are amino acid positions in Msb2. (C) Induction of the Hog1-specific reporter gene 8×CRE-lacZ by the hyperactive Msb2Δ1 mutant. TM257 was cotransformed with the reporter plasmid pRS414-8×CRE-lacZ and another plasmid carrying the galactose-inducible Msb2Δ1-2×HA (or its indicated mutants). The cells were grown in CARaf medium, and expression of Msb2Δ1-2×HA was induced by 2% galactose for 2 h. The activity of β-galactosidase in cell extracts was normalized to the cell density and is expressed as Miller units (35). Error bars represent SDs (n = 4). (D) Accessibility of the cysteine residues in Msb2 Ser/Thr-to-Cys mutants to maleimide-PEG2-biotin. TM257 was transformed with a plasmid that expresses Msb2Δ2-2×HA without other mutations (no mut.) or with the indicated mutations under the control of the GAL1 promoter. Expression was induced by 2% galactose for 2 h. Labeling and detection of cysteines accessible to 0.2 mM maleimide-PEG2-biotin were conducted essentially as described in the legend to Fig. 4B. (E and F) Direct disulfide bonding between Opy2 and Msb2. TM257 was cotransformed with a plasmid that expresses the indicated mutants of Opy2ΔSR1-Myc together with another plasmid that expresses the indicated mutants of Msb2Δ2-HA (E) or of Msb2Δ1-2×HA (F). Expression of these proteins is under the control of the GAL1 promoter. The cells were grown in CARaf medium, and expression of the tagged Opy2 and Msb2 was induced by 2% galactose for 2 h. Cell lysates were prepared using buffer A containing 0.2% Triton X-100 with (+) or without (−) 2 mM DTT. HA-tagged Msb2 was precipitated from cell lysates using an anti-HA antibody, and coprecipitated Opy2ΔSR1-Myc was probed with an anti-Myc antibody. Total expression levels of the tagged Opy2 and Msb2 proteins were uniform among the samples but are not shown in panel F to save space. (G) Schematic representation of the direct Opy2-Msb2 interaction. The numbers in the green boxes indicate amino acid positions in Msb2. Blue lines indicate disulfide bonding between Msb2 and Opy2. Other symbols are as described in the legend to Fig. 5E.