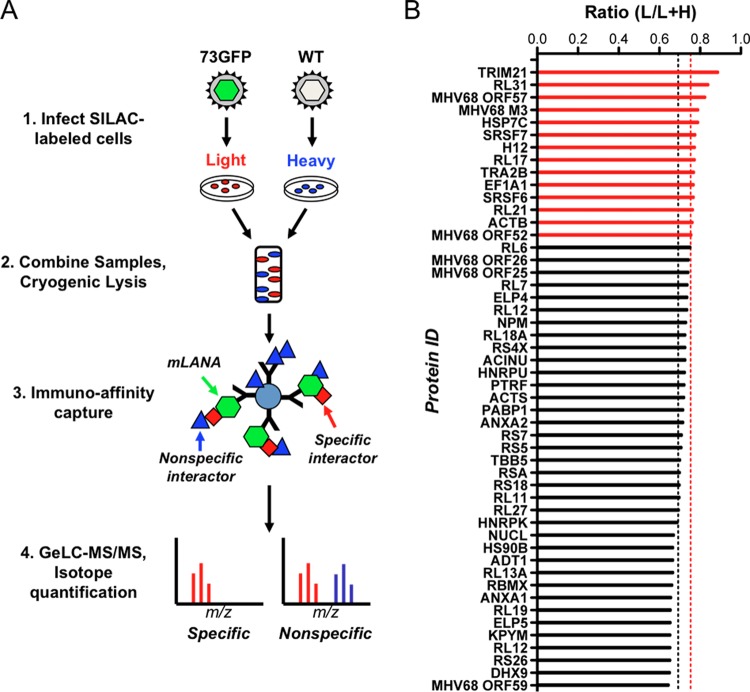
FIG 1.
Application of I-DIRT to identify mLANA-binding proteins in lytic MHV68 infection. (A) Overview of the I-DIRT method. Light or heavy isotopically labeled 3T3 fibroblasts were infected with either recombinant MHV68 expressing mLANA-GFP (73.GFP) or wild-type MHV68 (WT) at an MOI of 5 PFU/cell. At 18 h postinfection, intact cells were mixed at an equivalent ratio by weight, cryogenically disrupted, and resuspended in lysis buffer. mLANA-GFP and its interactors were immunoprecipitated using GFP antiserum covalently coupled to magnetic beads and purified for mass spectrometry. Isotopically light and heavy peptide fractions for a given protein interaction were calculated and scored as specific or nonspecific interactions based on a ratio calculated for common nonspecific protein contaminants. (B) mLANA-interacting proteins with both light and heavy peptides were classified as specific or nonspecific by I-DIRT. The fraction of light isotopic peptides containing [6-12C]arginine and [6-12C]lysine was calculated by dividing the peak area of the light peptide spectrum of a given protein by the sum of the peak areas for light and heavy peptide spectra for the same protein (ratio light/[light plus heavy]). The nonspecific threshold (0.6918 L/L+H, black dashed line) was determined by calculating the mean light fraction of ribosomal proteins that are common contaminants in interaction screens. Proteins with a light isotopic fraction greater than one standard deviation from the mean ribosomal threshold (>0.7528 L/L+H, dashed red line) are scored as specific interactions (red bars), while those with a fraction less than one standard deviation from the mean are classified as nonspecific interactions (black bars).