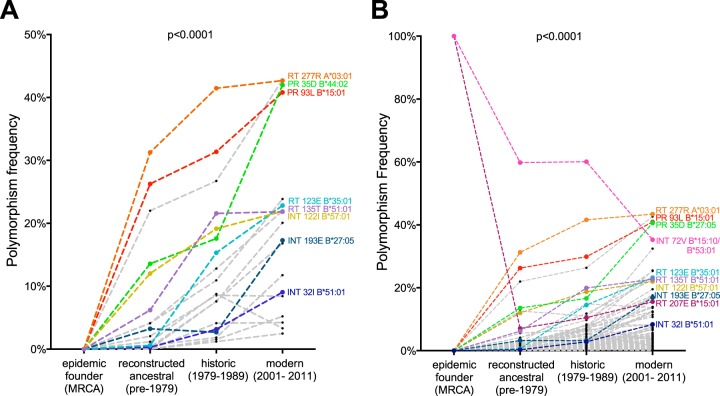
FIG 5.
Spread of HLA-associated polymorphisms evident from the earliest days of the North American epidemic. (A) Frequencies of nonconsensus conservatively defined HLA-associated polymorphisms in the inferred MRCA sequence, reconstructed ancestral sequences (1965 to 1979), historic sequences (1979 to 1989), and modern sequences (2001 to 2011). Polymorphisms of specific interest are labeled. (B) Frequencies of 120 comprehensively defined HLA-associated polymorphisms in the inferred MRCA sequence, reconstructed ancestral sequences, historic sequences, and modern sequences. Polymorphisms of specific interest are labeled. Polymorphisms RT 207E (associated with B*15:01) and INT 72V (associated with both B*15:10 and B*53:01) are present in the reconstructed MRCA sequence and decline in prevalence at the population level thereafter. Both analyses were agnostic to the individuals' HLA status, as the information was not available for the MRCA or reconstructed ancestors. The P value (P < 0.0001) for overall comparison was determined using the Friedman test. Historic and modern patients with known/suspected early infection were excluded from the analyses.