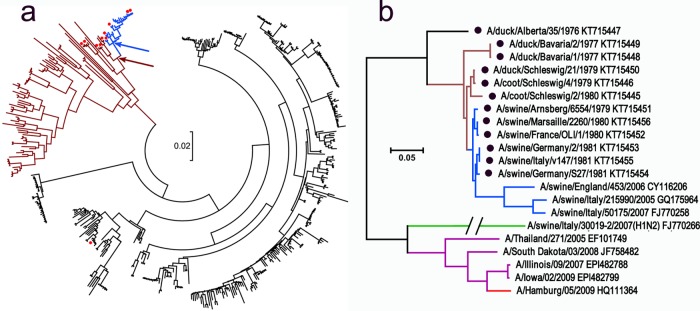
FIG 1.
Phylogenetic relationships between HAs of H1 influenza viruses. (a) Tree based on the sequences of 455 avian viruses from North America (black), Eurasia, Oceania, and Africa (brown) and avian-like swine viruses isolated in 1979 to 1981 in Europe (blue). Red dots depict virus strains that were tested in this study. Arrows show locations of hypothetical first swine virus (blue) and its putative avian precursor (brown). (b) Tree showing all of the viruses with H1 HA used in this study. Color coding of HA lineages: black, brown, and blue (same as in panel a); green, H1N2 swine viruses with human-like HA; purple, viruses with classical swine HA isolated from humans; red, H1N1/2009 pandemic virus. GenBank accession numbers are shown next to strain names. Two sequences (A/Illinois/09/2007 and A/Iowa/02/2009) (44) were obtained from GISAID EpiFlu Database (www.platform.gisaid.org/). Black dots depict sequences determined in the present study. The scale bars in both panels represent units of nucleotide substitutions per site.