FIG 3.
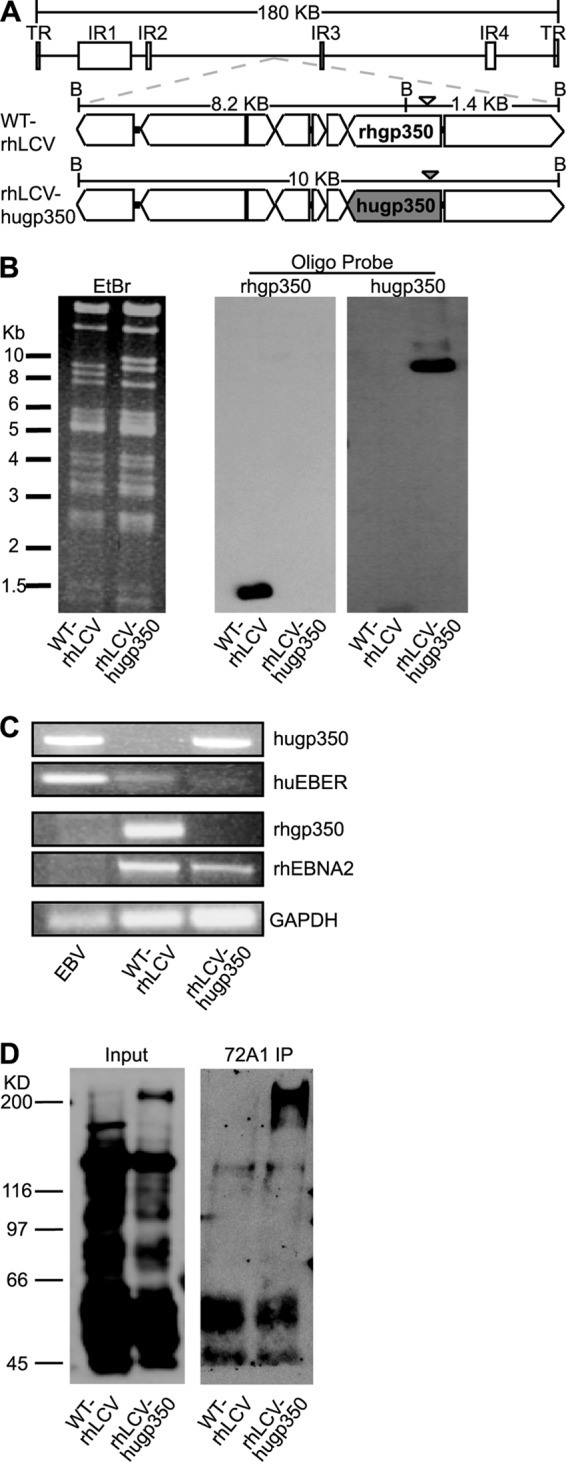
Generation of chimeric rhLCV-hugp350. (A) Schematic representation of the 10-kb region of the WT-rhLCV BAC (top) containing the gp350 open reading frame and surrounding genes. The native rhgp350 of the WT-rhLCV BAC is shown in the middle structure, and the replacement with the EBV gp350 in the chimeric rhLCV-hugp350 BAC is shaded in gray in the bottom structure. BamHI restriction sites are indicated by a B, and the distance between the restriction sites is noted. DNA probes used for detection of rhgp350 or hugp350 fragments after Southern blotting are shown as an open or filled arrowhead, respectively. (B) Validation of rhgp350 replacement with hugp350 in the rhLCV BAC. BamHI digests of WT-rhLCV BAC and chimeric rhLCV-hugp350 BAC recovered from the rhLCV-hugp350 LCL were separated by agarose gel electrophoresis. The ethidium bromide (EtBr)-stained gel is shown on the left. After Southern blotting, the membrane was hybridized with rhgp350- or hugp350-specific probes, as shown on the right. (C) The rhLCV-hugp350 LCL was infected with the BAC-derived chimeric rhLCV-hugp350. DNA from LCLs stably transformed with EBV, WT-rhLCV, and the chimeric rhLCV-hugp350 was assayed for hugp350, huEBER, rhgp350, and rhEBNA2 genes by PCR. GAPDH was amplified as a control. (D) The rhLCV-hugp350 LCL expresses EBV gp350. WT-rhLCV and rhLCV-hugp350 LCLs were induced for lytic replication, and EBV gp350 was precipitated from the cell lysates by incubation with 72A1. Whole-cell lysates are shown on the left, and 72A1 immunoprecipitation (IP) is shown on the right. EBV-immune human serum was used for immunoblotting.
