Fig. 7.
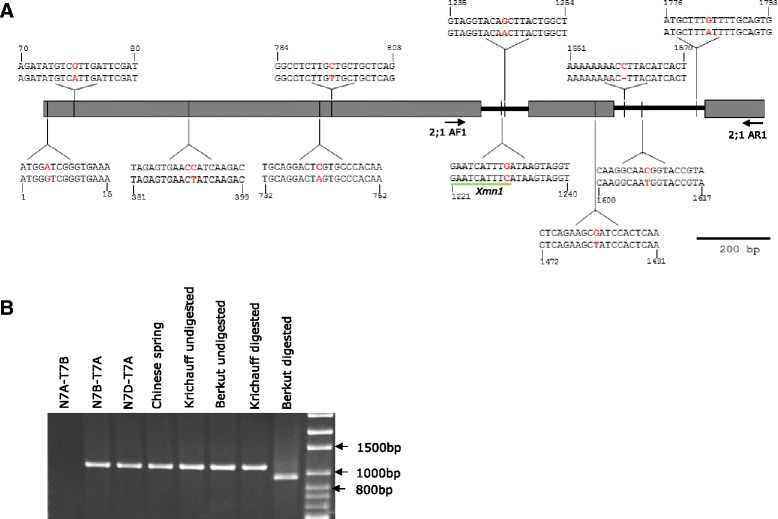
DNA sequence variability in the TaHKT2;1 7AL-1 gene between wheat cultivars Krichauff and Berkut. a Intron-exon structure where exons are represented as grey boxes and introns represented by black lines. SNPs are indicated by black vertical lines. DNA sequence and position (in base pairs) flanking each SNP is shown, with top sequence representing Krichauff and bottom sequence representing Berkut. SNP variation within the restriction enzyme recognition site, Xmn1, is underlined. Location of gene specific PCR primer pair for the TaHKT2;1 7AL-1, (2;1 AF1 and 2;1 AR1) to amplify the SNP at 1230 base pairs is shown by black arrows. b Agarose gel electrophoresis of TaHKT2;1 7AL-1 gene specific CAPS marker showing specificity to chromosome 7A using NT analysis and size difference of amplicons for Berkut and Krichauff parents following digestion with XmnI. The DNA ladder is shown to the right of the figure
