Figure 1.
ArrayEdit: an Arrayed, High-Content Platform to Monitor and Isolate Gene-Edited Cells via One-Pot Transcribed Single-Guide RNAs
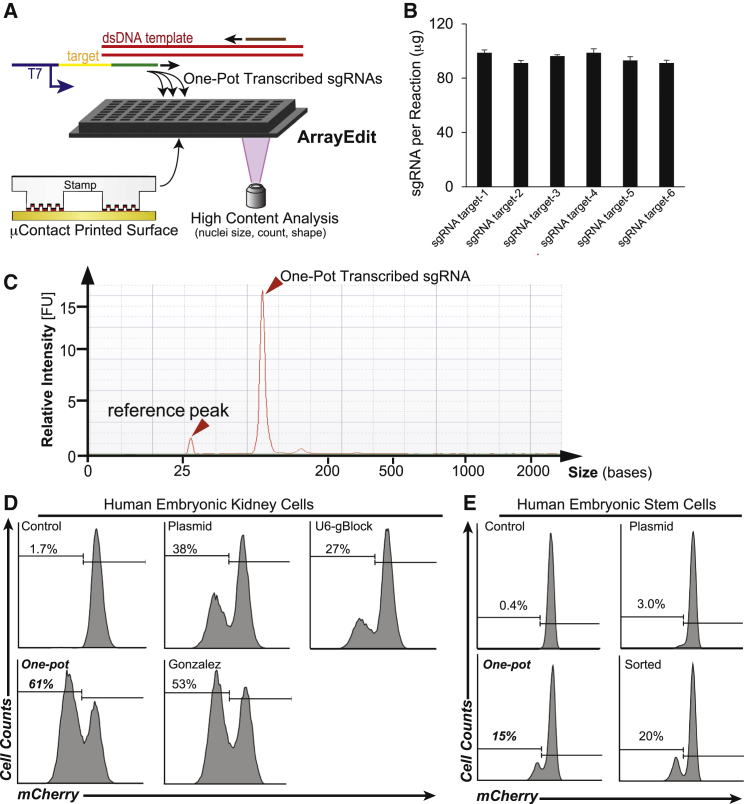
(A) Overview of ArrayEdit assembly and key components. Top: Schematic of one-pot PCR and T7 transcription. All components can be mixed and reacted within a single well without any intermediate purification steps. Primers are synthesized as custom oligonucleotides. The forward primer defines the genomic target of editing by Cas9. Bottom: Surface modification to the bottom of multiwell plates generates cell-adhesive μFeatures on a glass bottom. Each μFeature can be tracked over time via high-content imaging and stitched together to form a time-lapse visualization of edited cell phenotypes.
(B) Amount of sgRNA produced within each well via one-pot transcription. Data are represented as means ± 95% CI from four independent one-pot transcriptions on each sgRNA target (targets 1–3, mCherry-1-3; 4–6, GFP-1-3) and are not significantly different (Student's t-test, p > 0.05, Bonferroni correction).
(C) RNA Bioanalyzer spectra of one-pot transcribed sgRNA. Narrow peak (arrowhead) is consistent with only the desired product being produced. The reference peak is used by the Bioanalyzer to standardize size measurements.
(D and E) Flow cytometry histograms of HEK-H2B-mCherry cells (D) and WA09-H2B-mCherry hESCs (E) 4 days after delivery of mCherry-1 sgRNAs. sgRNA was either expressed of a plasmid, ordered commercially (U6-gBlock), created using previously described methods (Gonzalez), or one-pot transcribed (n = 2; independent experiments). Sorted: a GFP plasmid is co-electroporated and then sorted for GFP+ cells, leading to enrichment of cells that contained exogenous nucleic acids. See also Figure S1 and Tables S1 and S2.