Figure 2.
Deep Sequencing Reveals High-Efficiency Modification by One-Pot Transcribed sgRNAs
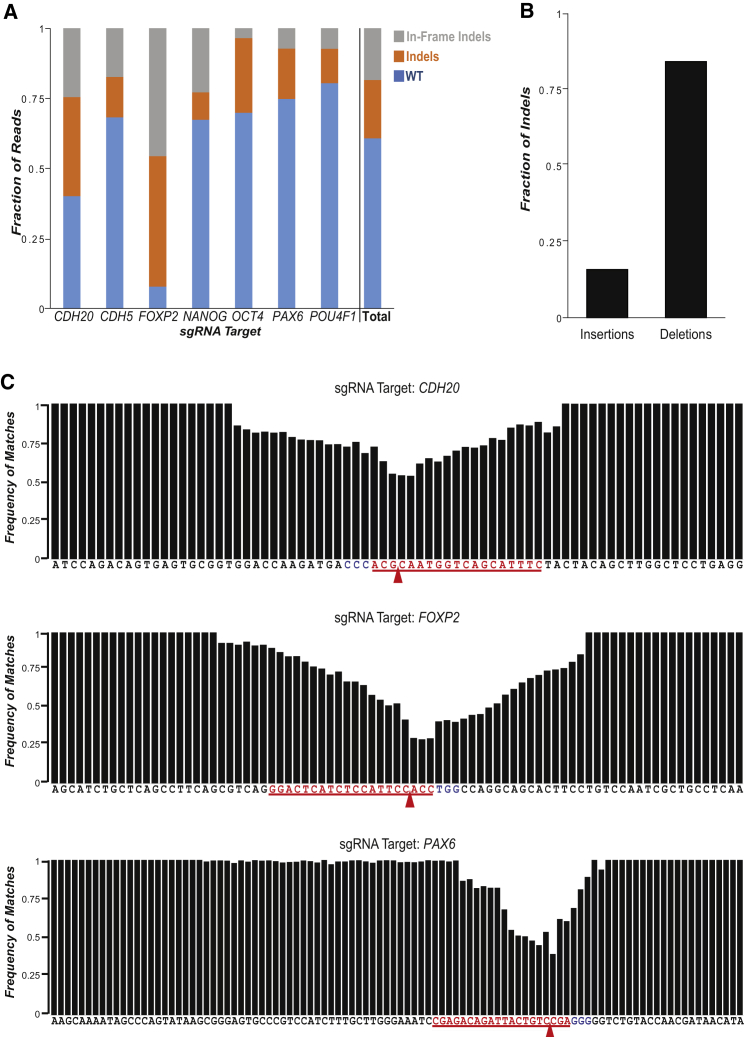
(A) Fraction of sequencing reads from gene-edited, hESC-derived embryoid bodies that matched the wild-type (WT) sequence (blue), contained at least one insertion or deletion (indel, orange), or contained an in-frame indel (gray). Edits occurred in genes and transcripts marking all three germs layers (ectoderm [PAX6, POU4F1], mesoderm [CDH5], endoderm [CDH20, FOXP2]) and pluripotent stem cells (NANOG, OCT4). Total: all reads across the seven loci.
(B) Frequency of insertion events to deletion events in the sequencing reads, with deletion being ∼5× more common than insertion.
(C) Per base frequency of matches to the wild-type sequence for three genes (CDH20, FOXP2, PAX6). Red bases denote the sgRNA target sequence while blue bases denote the protospacer adjacent motif (PAM). Red arrows indicate the predicted site of double-strand break formation by Cas9. The beginning and ending sequences (17–22 bp) for each gene are uniform, because they contain reads that were amplified using primers (17–22 bp) during PCR.