Figure 3.
Substrate Micropatterning Enables Live HCA of Gene Editing
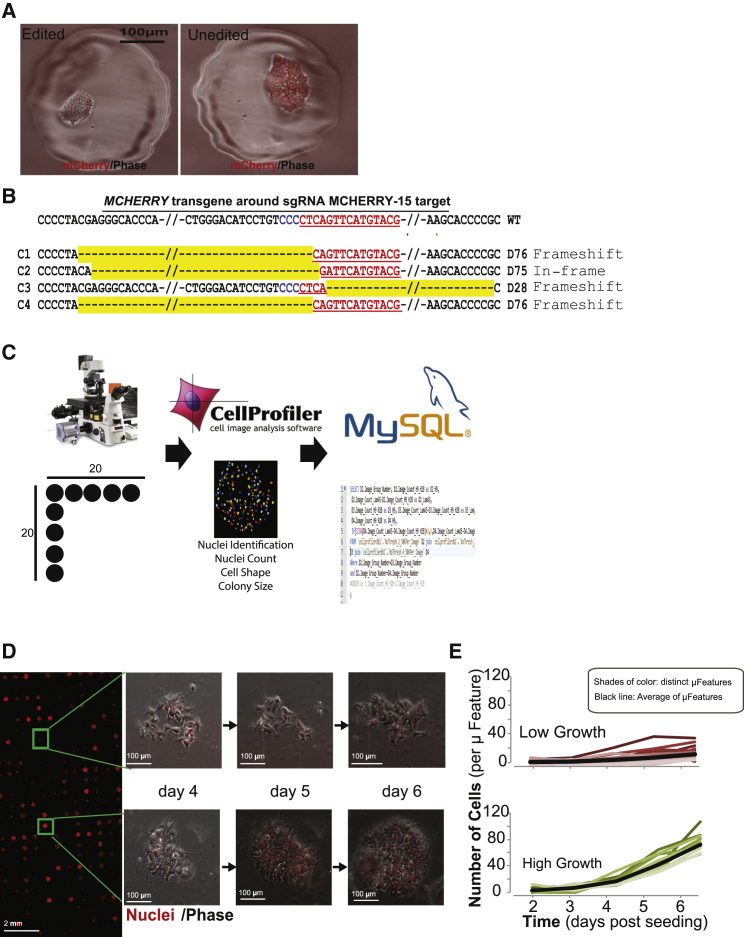
(A) Isolation of homogeneous mCherry gene-edited hESC lines on micropatterned plates. Clonal knockouts can be reliably identified and expanded in spatial isolation on each μFeature.
(B) Sanger sequencing of mCherry-edited clones isolated via ArrayEdit. The sgRNA target is denoted in red and the protospacer adjacent motif (PAM) in blue. Deletions are represented in yellow and the total length of deletion is to the right of the sequence (e.g., D76 indicates a deletion of 76 base pairs).
(C) High-content image acquisition and analysis workflow. Images are taken in a 20 × 20 grid and are passed to CellProfiler. Different colors indicate distinct identified objects (nuclei). CellProfiler results are sent to a MySQL database.
(D) Image of ArrayEdit within one standard culture well. Each μFeature can be tracked over time and stitched together to form a time-lapse visualization of edited cell phenotypes. Clones in two separate features are shown on days 4, 5, and 6.
(E) Growth curves for cells within 24 μFeatures on ArrayEdit over 5 consecutive days after editing with LAMA5 sgRNAs. Curves were separated into high- and low-growth rate groups. See also Table S3.