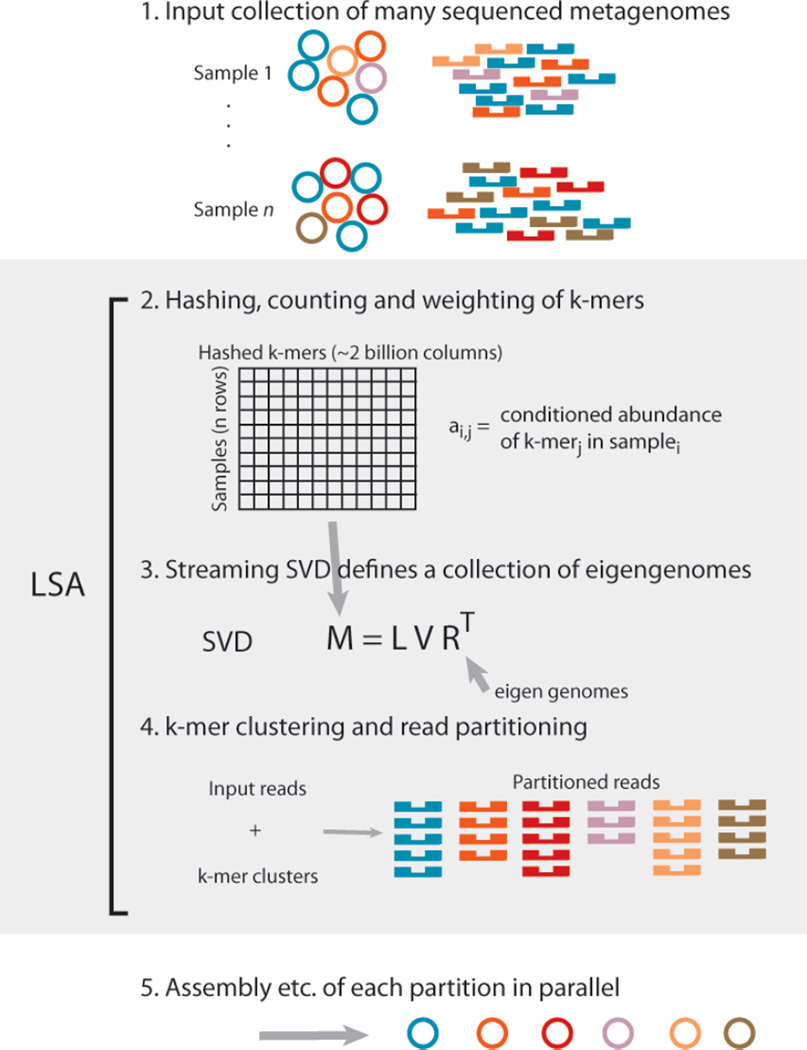
Figure 3.
Latent Semantic Analysis pipeline. Metagenomic samples containing multiple species (depicted by different colors) are sequenced. Every k-mer in every sequencing read is hashed to one column of a matrix. Values from each sample occupy a different row. Singular value decomposition of this k-mer abundance matrix defines a set of eigengenomes. K-mers are clustered across eigengenomes, and each read is partitioned based on the intersection of it’s k-mers with each of these clusters. Each partition contains a small fraction of the original data, and can be analyzed independent of all others.