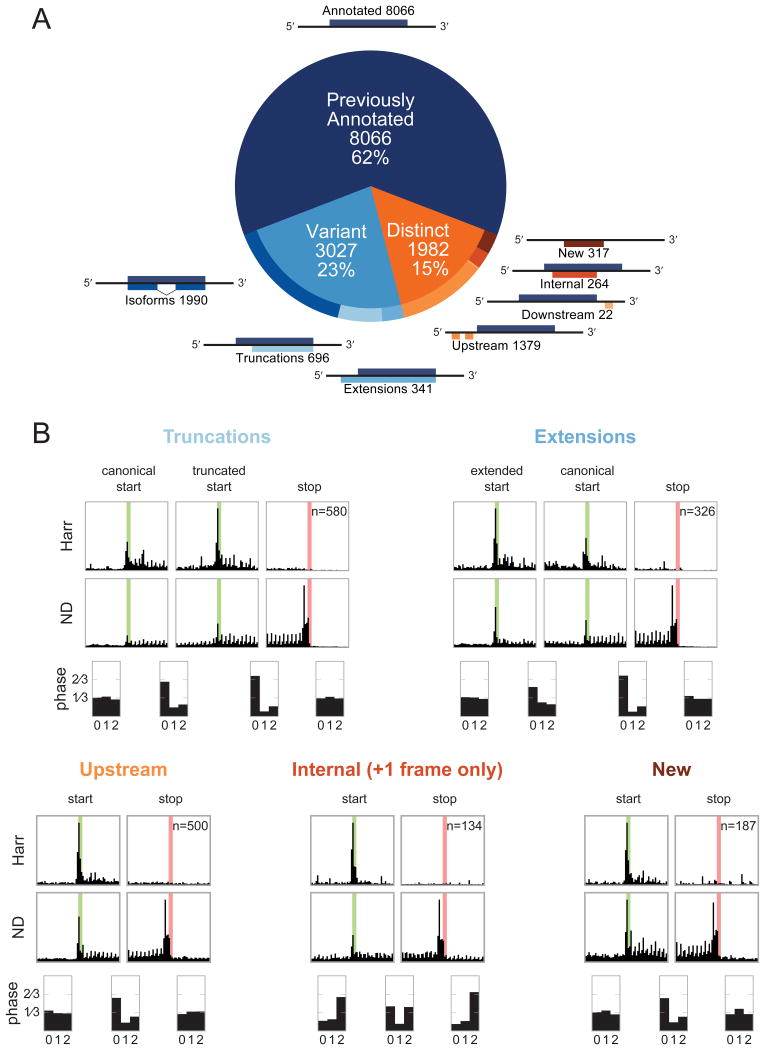
Figure 2. Previously unannotated translated CDSs in BMDCs fall into several classes, each of which displays patterns consistent with active translation.
(A) ORF-RATER identifies 13,075 high-confidence translated ORFs. The majority of these are previously annotated CDSs, and the majority of the remainder are variants of canonical CDSs that share portions of the coding sequence. ORFs distinct from annotated CDSs occur primarily in 5′ UTRs, though a sizable subset are found on transcripts without previously appreciated coding potential or in alternate frames of canonical CDSs. See Figure S1A for the distribution of ORF-RATER scores for each type, and Table S1 for a complete list of all high-confidence CDSs. (B) Metagene profiles of each class of new CDS display the hallmarks of translation, including peaks of density at newly identified start codons following Harr treatment, peaks of density at stop codons under ND treatment, and greater read density in between. Translated truncations (top left) and extensions (top right) display peaks of density at both the canonical and novel translation initiation sites, suggesting that both are used on average. The average read density in all translated regions show 3-nucleotide periodicity in the expected reading frame, with the exception of internal CDSs, for which the reading frame is on average a superposition of the canonical and alternative frames. Metagene profiles for the LTM and CHX datasets are plotted in Figure S2.