Abstract
When seeking to reproduce results derived from whole-exome or genome sequencing data that could advance precision medicine, the time and expense required to produce a patient cohort make data repurposing an attractive option. The first step in repurposing is setting some quality baseline for the data so that conclusions are not spurious. This is difficult because there can be variations in quality from center to center, clinic to clinic and even patient to patient. Here, we assessed the quality of the whole-exome germline mutations of TCGA cancer patients using patterns of nucleotide substitution and negative selection against impactful mutations. We estimated the fraction of false positive variant calls for each exome with respect to two gold standard germline exomes, and found large variability in the quality of SNV calls between samples, cancer subtypes, and institutions. We then demonstrated how variant features, such as the average base quality for reads supporting an allele, can be used to identify sample-specific filtering parameters to optimize the removal of false positive calls. We concluded that while these germlines have many potential applications to precision medicine, users should assess the quality of the available exome data prior to use and perform additional filtering steps.
1. Introduction
Although the costs of whole-exome sequencing continue to decrease [1], the resources needed to identify, enroll, and sequence an entire cohort of interest will remain significant for the foreseeable future. This process is especially cumbersome when investigating rare phenotypes, including certain cancers and tumor subtypes. A more convenient alternative path is to identify and then repurpose publicly accessible datasets in order to test new hypotheses or to reproduce findings of studies performed on independent cohorts. Federal policies explicitly promote data sharing and repurposing, by supporting public repositories like the database of Genotypes and Phenotype (dbGaP) and the Sequence Read Archive (SRA) [2,3]. The challenge, however, is that diverse datasets each developed with different goals in mind will often have unique features that require special care before they can be pooled together for repurposing. Clearly, the quality of exome variant calls varies by platform and depth of the sequencing [4,5] and also depends on the stringency of downstream pipelines for SNV identification and variant filtering [6]. Currently, most whole-exome quality assessment tools focus on evaluating the quality of the raw input data [7,8] rather than on the output calls; moreover, approaches that do assess the output generally limit themselves to comparing calls to 1000 Genomes Project or dbSNP variants [9,10] without providing recommendations for filtering or even clear conclusions on whether the data is acceptable for use. Yet, if a dataset is repurposed inappropriately, systematic biases and variability in noise levels may slant results, lower reproducibility, yield artifacts, or prevent confirmation of prior findings [11]. This presents a major problem for precision medicine in particular, since targeting a falsely called variant may result in ineffective treatment.
In order to probe the impact that dataset and variant filtering choices can have on the quality of repurposed data, we assessed in detail germline exomes from The Cancer Genome Atlas (TCGA) [12]. TCGA currently gathers diverse information from more than 11,000 patient samples across 34 cancer types. Final germline variant calls for some cancer types are available through the TCGA Data Portal, with additional lower level sequence data also available from the CGHub repository (https://cghub.ucsc.edu/). However, the primary goal of sequencing cancer patient germline samples was to provide the background information that will enable the recognition of somatic variants unique to the tumor. Secondary use of these germline exomes to further precision medicine has thus far been uncommon but shows the promise of using these germlines to predict response to treatment within a cancer cohort, detect genetic differences in individuals who develop cancer, and identify germline contributions to the process of tumorigenesis [13,14,15].
Here, we evaluated the quality of TCGA germline single nucleotide variation (SNV) calls in a given exome by testing whether two features of their collected variant calls followed the known biology of substitution and purifying selection or whether these features were lost and suggested that the variant calls were of non-biological origin.
The first feature, called Ti/Tv, has been previously described and is based on the biology of spontaneous base substitutions. In the germline, these are more often transitions (from purine to purine, or from pyrimidine to pyrimidine) than transversions (from purine to pyrimidine or pyrimidine to purine) so the Ti/Tv ratio is normally >3 across an exome, whereas for random base changes as one might produce computationally Ti/Tv is equal to 0.5 [10]; this difference can then serve as a proxy for germline variant call quality [10,16,17].
The second and novel feature, called λ, is based on the biology of purifying selection of germline mutations. Fisher’s geometric model [18] predicted in 1930 that the distribution of fitness effect of germline mutations would follow a decaying exponential. For the first time, a recent study of the Evolutionary Action (EA) of human polymorphisms [19] provides a measure for the fitness effect of mutations and for their negative selection, through a decay constant λ, which is much larger for biological germline mutations than for random mutations.
Therefore, we hypothesized that Ti/Tv and λ related to the substitution and selection processes, respectively, should be complementary measures of variant call quality. Our findings support this view and also emphasize the importance of using multiple, orthogonal quality measures. Using these measures we estimate the fraction of false positive variant calls in TCGA exomes and reveal substantial variability in quality by sample, cancer subtype, and sequencing source. The methods described in this study provide an easy way to assess the quality of germline exome data and suggest, for the first time to our knowledge, the importance and feasibility of sample-specific filtering parameters.
2. Methods
2.1. Exome data acquisition
Gold standard germline exome variant callsets were obtained from http://www.illumina.com/platinumgenomes/[20]. The NA12877 and NA12878, v7 (released December 2014) merged callsets were downloaded, and only ‘platinum’ calls limited to those with a ‘PASS’ designation in the filter field were considered confident calls in subsequent analysis. 1000 Genomes Project [21] Phase 3 exomes were downloaded from http://www.1000genomes.org/ on 5/11/15. All cancer germline exomes available on 10/23/14 were downloaded from TCGA [12] and separated by cancer type and institution.
2.2. Quality Assessment
The Ti/Tv ratios were calculated for each exome variant callset by counting all coding purine to purine and pyrimidine to pyrimidine (transition) SNV mutations, and dividing this value by the number of purine to pyrimidine and pyrimidine to purine (transversion) SNV mutations.
To measure the selection decay λ of variant callsets, the corresponding vcf files were annotated with ANNOVAR [22] and the Evolutionary Action (EA) was computed to measure the fitness effect of all missense mutations. Histograms of the distribution of EA scores, binned by deciles, were fitted to an exponential curve (Eq. 1) using a least-squares-fit to estimate λ. In Eq. 1 the ‘x’ values represent Evolutionary Action, ‘y’ represents proportion of mutations, and A and λ are constants.
| (1) |
2.3. Simulated Variant Callsets
Both Ti/Tv and λ were calculated for 100 datasets with different fractions of computer-generated false positive variant calls. The fraction varied from 0 to 1, in increments of 0.01. Each dataset consisted of 1000 simulated exome files, each of which contained a total of 10000 missense mutations. The mutations were drawn either from the ‘true positive’ pool of variants from gold standards NA12877 and NA12878, or randomly from the ‘false positive’ pool of all possible human SNVs in order to create the proper ratio. The average and standard deviations of λ and Ti/Tv for each fraction of false positives were calculated, and their correlations with percent noise fitted with exponential.
2.4. Application of λ to TCGA cohorts
For each of 21 cancer types, all germline datasets were separated by institutional source, curation, and sequencing platform. When multiple variant callsets were available for a given cancer type, files marked ‘curated’ were chosen over those which were not marked curated. If multiple curated sets were available, λ values were calculated for each and the set with the highest average λ was chosen. If no curation was noted on any of the files, λ values were calculated for all available sets and the set with the highest average λ was chosen. For each sample, the predicted percentage of false positive calls was then calculated from Eq. 2.
3. Results
3.1. Calculating quality measures for ‘gold standard’ germline exomes
First, we calculated the quality scores Ti/Tv and λ (Figure 1) for gold standard germline exome calls: Illumina Platinum v7.0 variant calls for samples NA12877 and NA12878 [9]. These samples were sequenced to 200× depth on a HiSeq 2000 system, and 15 other members of their pedigree were sequenced to 50× depth on the same system. Variant calls deemed ‘platinum’ take into account inheritance constraints in the pedigree as well as concordance of variant calls across multiple aligners and callers [24,25,26,27,16]. As described in Methods, the transition to transversion ratio (Ti/Tv) detects a biological bias in the types of mutations that occur in the call set, while the λ shows the purifying pressure against coding mutations as a function of their Evolutionary Action (EA) [19]. For the platinum whole-exome calls for NA12877 and NA12878, we found the Ti/Tv ratios to be 3.05 and 3.04 for whole-exome variant calls and 1.97 and 1.96 for missense variant calls respectively, while λ was 0.0379 and 0.0380 respectively. These values were also reasonably consistent with exome data from The 1000 Genomes Project [21] and were independent of ethnic background; the average λ is 0.0379±0.0008 and the average Ti/Tv is 3.11±0.05 for whole-exome variants and 2.06±0.04 for missense variants. These data show a gold standard ‘target’ for these quality parameters that were used to assess other datasets.
Fig. 1.
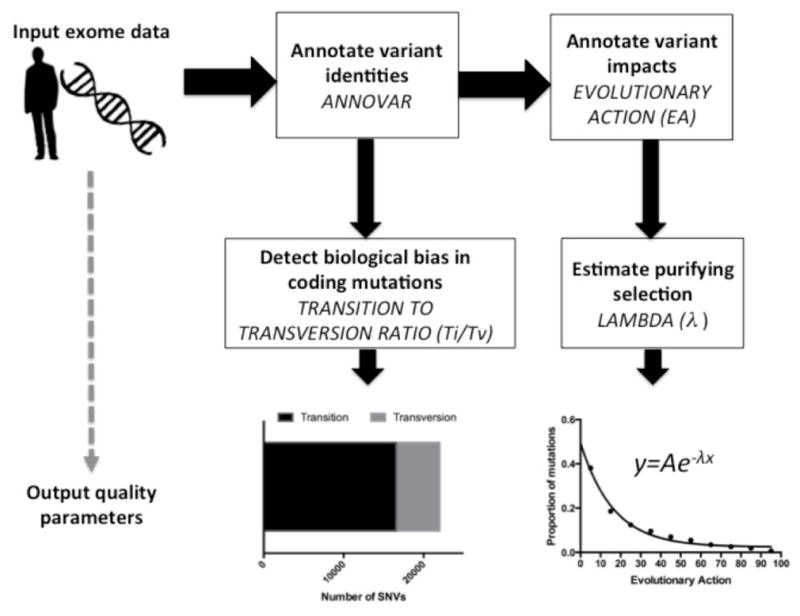
Illustration depicting the steps taken to calculate λ and Ti/Tv parameters from exome data.
3.2. Quality measures λ and Ti/Tv decrease in a predictable fashion as false positives are added to a variant call set
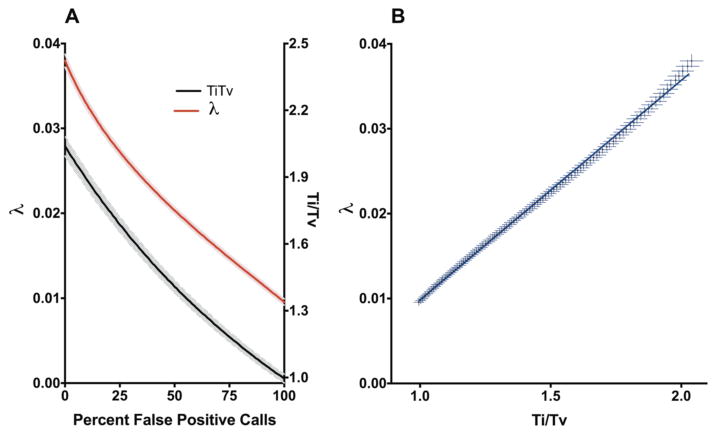
We next calculated the λ and Ti/Tv for SNV sets with varying fractions of false positive calls. Using the Illumina Platinum calls as a pool of true positives (TP) and all possible human SNVs generated by random nucleotide changes as a pool of false positives (FP), 1000 simulations of 10000 missense mutations were produced for each of 100 TP:FP ratios. As the fraction of the variant calls composed of random noise increases, both λ and Ti/Tv decrease in an exponential fashion (R2 > 0.99) such that
| (2) |
(Figure 2A). In addition, the two quality measurements were strongly correlated with one another (Pearson R = 0.9993, p < 0.0001) (Figure 2B). Use of this simulated data enables us to estimate, for a set of real exome variant calls, the degree of contamination by false positives in the sample.
Fig. 2.
Simulated noise in exome SNV calls. (a) Effect of increased noise on λ and Ti/Tv values. Shaded regions indicate the standard deviation around the mean. (b) Correlation between λ and Ti/TV.
3.3. TCGA germline datasets vary in the amount of noise they contain
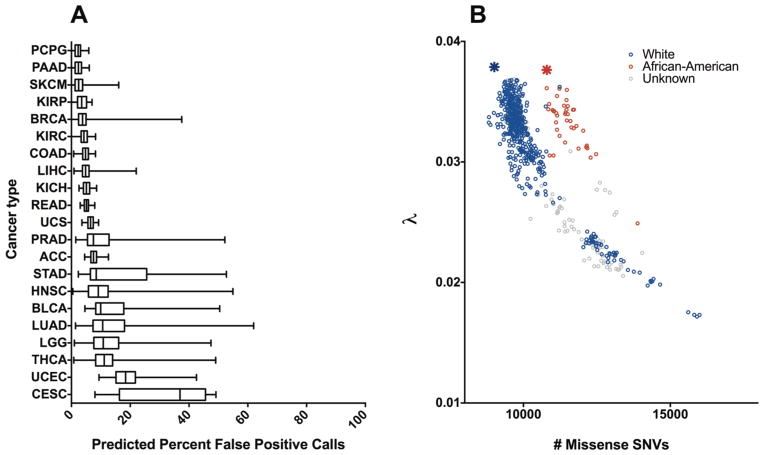
Next, we estimated the fraction of false positive variant calls in whole-exome germline data from TCGA and assessed whether the fraction is consistent within each cancer subtype. Germline variants were available through TCGA for 21 cancer types, where they were organized by sequencing institution, sequencing platform, and curation level. Some tumors were sequenced by multiple institutes, though the extent of this overlap depended on the cancer type. We focused on data marked ‘curated’, rather than those marked ‘automated’, and when there were still multiple versions we chose the (presumably best) dataset with the largest λ (see Methods). The predicted percent of false positive calls for each exome was calculated based on Eq. 2, and the distribution for each cancer type is shown in Figure 3A. At the extremes, pheochromocytoma and paraganglioma (PCPG) exomes were predicted to uniformly contain less than 5% false calls, while most cervical cancer (CESC) exomes were predicted to contain more than 35% false calls. The datasets also differed greatly in variance, with rectal adenocarcinoma (READ) exomes having less than 10%, and lung adenocarcinoma (LUAD) having more than 60% variances. These results do not necessarily reflect intrinsic differences between cancer types. Large numbers of variant calls in an exome corresponded to lower λ scores and indicated an excess of false positives, as shown for LUAD in Figure 3B. However, using the number of mutations as a proxy for the false positive rate may be misleading when the cohort consists of individuals with diverse ethnic backgrounds; for example, exomes from patients of African ancestry consistently had more variants when compared to other exomes with the same λ values (Figure 3B), in agreement with data from the 1000 Genomes Project. The estimated number of ‘true’ missense SNV calls was consistent between samples of the same ethnic background and fit the numbers of missense mutations seen in the 1000 Genomes cohorts. These data show the marked heterogeneity of false variant call rates in TCGA germline exomes and highlight the hazards of using these datasets as-is.
Fig. 3.
Application of λ to TCGA cohorts. (a) Predicted noise across 21 TCGA cancer types. The data are represented in a box-and-whiskers plot that uses the center line to indicate median, the box to indicate quartiles, and the whiskers to indicate range. Cancer types are ordered by median. (b) Exponential relationship between λ and number of missense SNVs in Lung Adenocarcinoma. Associated open-access clinical data provided by TCGA was used to separate patients by their self-identified race. The average lambda/number of missense mutations for the 1000 Genomes Project Caucasian (CEU) and African-American (ASW) cohorts are noted with a blue and red star, respectively.
3.4. Data quality is not consistent across calling centers
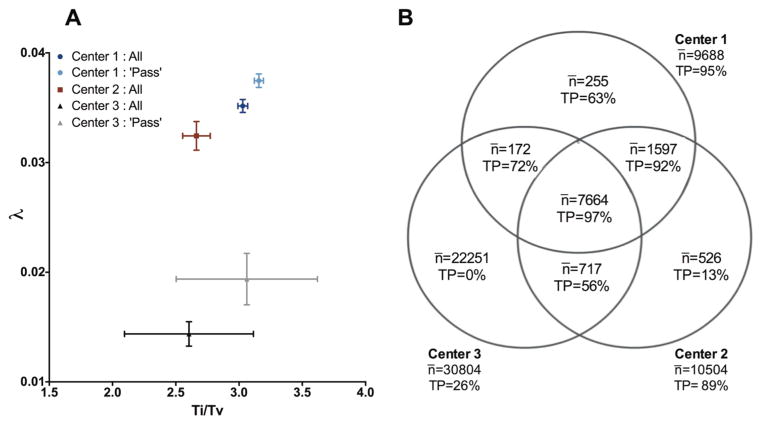
In order to test reproducibility of data across sequencing centers, we focused our analysis on the chromophobe renal cell carcinoma (KICH) dataset, which consisted of SNV germline calls from three separate institutions for the same patients. For each sequencing center, we calculated the average Ti/Tv ratio as well as the λ selection decay constant for the germline variants of each exome, as shown in Figure 4A. The center with the highest average Ti/Tv ratio also had the highest λ, which corresponded to an average of ~5% false positive calls per sample. For the second center, both the Ti/Tv ratio and λ were lower, and λ predicted an average of ~12% false positive calls per sample. For Center 3, although the average Ti/Tv ratio was nearly as good as the other two centers (2.6 compared to 2.66 and 3.1, respectively), the average λ was radically different (0.014 versus 0.032 and 0.036, respectively) and suggested that on average 76% of the calls in each sample were false positives. Indeed, the average number of missense SNV calls from this center (31,000±5,000) was over 3 times higher than the Illumina platinum exomes defined above as gold standard, which further supported a false positive rate of at least 70%. The different sensitivity of Ti/Tv and λ to lower quality variant calls in this case may be due may be due to technical aspects of the calling methods themselves; if the known biological bias toward transition mutations was built into the calling algorithm used by Center 3 and it was used as a factor in deciding whether to report a variant, even false positive mutations will have a high Ti/Tv. In this case, λ detected noise whereas Ti/Tv was equivocal, stressing the importance of using multiple quality measures in exome data assessment. For centers 1 and 3, additional internal filters separated the reported calls into those that ‘pass’ and those that do not. Restricting our analysis only to passing calls improved quality but was not sufficient to eliminate either the detected noise or the center-specific differences. These data show that the germline variant calls of TCGA patients made by different sequencing platforms and calling pipelines are sometimes very different and require careful examination by multiple, orthogonal quality measures.
Fig. 4.
KICH SNV calls for three centers (a) λ and Ti/Tv of calls. For each patient assessed by each center, λ and Ti/Tv were calculated and the average and standard deviations of these values are displayed by institution. For centers 1 and 3, internal ‘pass’ filters were available and are displayed as well. (b) Predicted percentage of true calls for calls agreed upon by 1, 2, or 3 institutions. For 65 KICH patients assessed by all three centers, all calls regardless of internal filtering were separated by the institution(s) that identified them. The average number of missense mutations per patient, as well as the predicted percentage of true positive calls derived from the λ value of the call set, is shown for each possible combination of sites.
A common practice to address such inconsistency in reproducibility is either to merge the available data or to use their intersection. In the first case, combining the calls from all centers would add substantial noise from Center 3. Using the intersection of all three centers, on the other hand, would result in high quality but roughly two thousand true positive calls per exome would be left out. Restricting to calls made by at least two of the centers may seem like a reasonable middle ground, but even this may not be the optimum solution. Figure 4B shows that calls made by Center 1 alone were still predicted to be of higher quality than calls agreed upon by Centers 2 and 3. These data demonstrate the caveats of ‘common sense’ filtering and highlight the importance of examining data quality carefully before integrating information from multiple sources.
3.5. Appropriate filtering parameters for SNV calls are sample-dependent
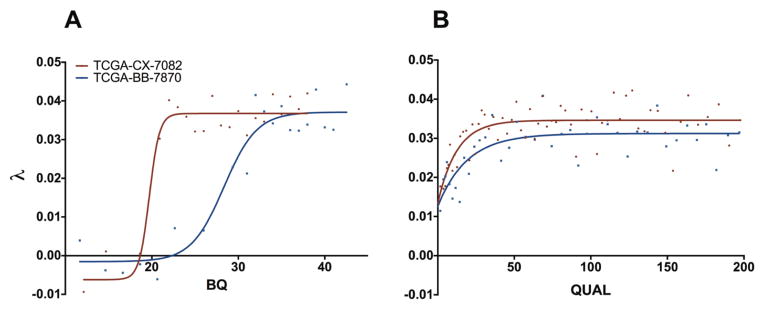
Having used λ to detect and quantify the presence of noise within these datasets, we next explored whether λ can be used for filtering false positive SNV calls. For illustration, we used exome data from two head and neck squamous cell carcinoma (HNSC) cancer patients. Each SNV in these data was associated with an average base quality for reads supporting alleles (BQ value) and a Phred-scaled quality score (QUAL value), amongst other features. For each patient, missense mutations were partitioned by BQ value and QUAL value in turn and each bin was assessed for λ. We found that λ depended on the BQ score with a sigmoidal relationship (R2 > 0.9), which indicated that below a BQ cutoff the SNV calls became random (Figure 5A). Strikingly, this cutoff is specific to each exome, even those sequenced on the same date, by the same center, on the same sequencing platform, and using the same SNV calling pipeline. For example, SNVs with a BQ of 25 appeared to retain high quality in one patient but to be comprised entirely of noise in the other (see blue curve in Fig. 5A). We also found that λ depended on the QUAL value of SNVs, such that the fraction of true positive calls was lower for QUAL values near zero and gradually increased with QUAL value till it reached a plateau at about QUAL=40 (Figure 5B). For the two exomes of Figure 5b the QUAL value did not correspond to the same fraction of false positive data, since the two exomes reached different maximum λ values. These sample-dependent differences suggested that BQ and QUAL values should only be interpreted as relative measures within the context of a given exome, and that filtering parameters should be customized for each exome using λ in order to achieve optimal separation of true and false positive calls.
Fig. 5.
Relationship between SNV features and λ for two HNSC patients. (a) Relationship between BQ and λ. For each patient, all missense SNVs were partitioned by BQ value such that every bin contained at least 50 calls; points represent the λ and average BQ of the bin. Solid lines represent sigmoidal fits. (b) Relationship between QUAL and λ. For each patient, all missense SNVs were partitioned by QUAL value such that every bin contained at least 50 calls; points represent the λ and average QUAL of the bin. Solid line represents fit to equation . For display purposes values of QUAL higher than 200 were not shown.
4. Discussion
Assessing the quality of genetic variant calls has great practical importance to precision medicine, since various sequencing platforms, coverage depths, and bioinformatics pipelines to call variants result in the inclusion of an unknown number of false positive calls. This becomes a major concern when naïve users access and repurpose publically available exome data assuming all reported calls are reliable. Here, we call attention to the hazards of this assumption by applying two measures of exome data quality, the transitions-to-transversions ratio and the purifying selection pressure (λ) of variant calls, to publically accessible data. As a test system we used germline exomes of 21 cancer types available through TCGA, which were generated with the primary purpose of being a reference for calling somatic mutations. We found considerable variation in data quality between and within cancer types, such that repurposing these data as-is may mislead scientists to conclude a lack of reproducibility and unsuccessful validation of previous findings, which would hinder the progress of precision medicine as a field.
As a gold standard of true germline variants we used the Illumina Platinum samples, NA12877 and NA12878, which are the current state-of-the-art high-confidence variant calls. However, this gold standard may still include some false calls, and future developments may allow for even more accurate sequencing and variant calling. These estimates, however, have great practical importance in comparing the calling confidence of two or more exomes. Indeed, the large differences in the average fraction of false positive calls between TCGA germline exomes of different cancer types, as well as the surprising variability within a cancer cohort, underscore the need to examine all data carefully before reuse in order to improve the reproducibility of results.
When we compared the germline variants called from three different sequencing centers for the same patients, we found a considerable lack in reproducibility between centers. However, classifying variant calls by their concordance across centers was revealing. Variants called by all three centers were assessed to contain only about 3% of false positive calls; in contrast, variants called by just one of the three centers had deviant λ values that matched the simulated introduction of up to 100%, 87% and 37% false positives. For each given center, unique variant calls were predicted to contain more false positives than those also called by at least one of the other centers; still, we found that depending on the relative data quality between centers even the unique variants of a single center may contain less noise than variants agreed upon by the other two centers. This calls into question the common practices of merging data or using the overlap of calls from different centers, which may include many false positive calls or exclude true positive calls, respectively. While it was useful to exclude variants that were not annotated with “PASS” in vcf files, this filter was not able remove all or even most false positive calls. Thus, the use of λ selection pressure analysis presents a rational, quantitative approach to determining which data should be used in association studies.
Features of the SNVs, such as quality scores, can also be used to filter out false positive calls. This basic principle is already established in post-processing variant calls, but many users apply ‘hard filters’ to all samples and express uncertainty regarding the appropriate filters to use. Using arbitrary cutoffs for all TCGA exomes without considering quality assessment will cause some samples to retain substantial numbers of false calls and others to lose many true calls. Our results suggested that BQ value produced the most effective separation between predicted true and false positives, and that the appropriate BQ value cutoff was different for each exome, even when the data were produced by identical procedures. This analysis allows users to leverage the relationship between λ and BQ in order to choose for each exome the optimum cutoff, allowing them to repurpose these datasets with confidence and improve the reproducibility of their results.
Assessing the quality of germline variant calls is a pressing issue both to improve their intended use as well as to facilitate their repurposing for secondary goals, and since an increasing amount of exome data is being deposited in public databases. Here, we show that elementary evolutionary considerations provide a general and simple approach to detect random sequencing errors. Whereas high quality data contain variant calls that follow an invariant and known distribution of Evolutionary Action, false positive variant calls recognizably distort this Action distribution. Remarkably, this distortion can classify sequenced genomes by quality and also separate variant calls by quality within single exomes on a case-by-case basis. This work reveals wide quality disparities in sequencing data but also demonstrates how this can be overcome through the use of the Evolutionary Action concept. In the future it should therefore be possible to apply, pool and repurpose public genome sequencing data with full confidence in their quality leading to better correlations with clinical phenotypes and enhancing reproducibility in precision medicine.
Acknowledgments
Funding: National Institutes of Health R01 grants GM079656-8, GM066099.
Footnotes
Conflict of interest: none declared.
Contributor Information
AMANDA KOIRE, Email: koire@bcm.edu, Program in Structural and Computational Biology and Molecular Biophysics, Baylor College of Medicine, Houston, Texas 77030, USA.
PANAGIOTIS KATSONIS, Email: katsonis@bcm.edu, Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, Texas 77030, USA.
OLIVIER LICHTARGE, Email: lichtarg@bcm.edu, Department of Molecular and Human Genetics Baylor College of Medicine, Houston, Texas 77030, USA.
References
- 1.Hayden EC. Nature. 2014;507(7492):294–5. doi: 10.1038/507294a. [DOI] [PubMed] [Google Scholar]
- 2.Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, Popova N, Pretel S, Ziyabari L, Lee M, Shao Y, Wang ZY, Sirotkin K, Ward M, Kholodov M, Zbicz K, Beck J, Kimelman M, Shevelev S, Preuss D, Yaschenko E, Graeff A, Ostell J, Sherry ST. Nat Genet. 2007;39(10):1181–6. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leinonen R, Sugawara H, Shumway M. Nucleic Acids Res. 39(Database issue):D19–D21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark MJ, Chen R, Karczewski KJ, Chen R, Euskirchen G, Butte AJ, Snyder M. Nat Biotechnol. 2011;29(10):908–14. doi: 10.1038/nbt.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meynert AM, Bicknell LS, Hurles ME, Jackson AP, Taylor MS. BMC Bioinformatics. 2013;14:195. doi: 10.1186/1471-2105-14-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo Y, Ye F, Sheng Q, Clark T, Samuels DC. Brief Bioinform. 2014;15(6):879–89. doi: 10.1093/bib/bbt069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pabinger S, Dander A, Fischer M, Snajder R, Sperk M, Efremova M, Krabichler B, Speicher MR, Zschocke J, Trajanoski Z. Brief Bioinform. 2014;15(2):256–78. doi: 10.1093/bib/bbs086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bao R, Huang L, Andrade J, Tan W, Kibbe WA, Jiang H, Feng G. Cancer Inform. 2014;13(Suppl 2):67–82. doi: 10.4137/CIN.S13779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heinrich V, Kamphans T, Stange J, Parkhomchuk D, Hecht J, Dickhaus T, Robinson PN, Krawitz PM. Genome Med. 2013;5(7):69. doi: 10.1186/gm473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Q, Guo Y, Li J, Zhang B, Shyr Y. BMC Genomics. 2012;13(Suppl 8):S8. doi: 10.1186/1471-2164-13-S8-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoff KJ. BMC Genomics. 2009;12(10):520. doi: 10.1186/1471-2164-10-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.TCGA Research Network. http://cancergenome.nih.gov/
- 13.Birkbak NJ, Kochupurakkal B, Izarzugaza JM, Eklund AC, Li Y, Liu J, Szallasi Z, Matulonis UA, Richardson AL, Iglehart JD, ZC PLoS One. 2013;8(11):e80023. doi: 10.1371/journal.pone.0080023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanchi KL, Johnson KJ, Lu C, McLellan MD, Leiserson MDM, Wendl MC, Zhang Q, Koboldt DC, Xie M, Kandoth C, McMichael JF, Wyczalkowski MA, Larson DE, Schmidt HK, Miller CA, Fulton RS, Spellman PT, Mardis ER, Druley TE, Graubert TA, Goodfellow PJ, Raphael BJ, Wilson RK, Ding L. Nature Communications. 2014;5:3156. doi: 10.1038/ncomms4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ngeow J, Ni Y, Tohme R, Song Chen F, Bebek G, Eng C. J Clin Endocrinol Metab. 2014;99(7):E1316–21. doi: 10.1210/jc.2014-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rimmer A, Phan H, Mathieson I, Igbal Z, Twigg SR, Wilkie AO, McVean G, Lunter G WGS500 Consortium. Nat Genet. 2014;46(8):912–8. doi: 10.1038/ng.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lam HY, Clark MJ, Chen R, Chen R, Natsoulis G, O’Huallachain M, Dewey FE, Habegger L, Ashley EA, Gerstein MB, Butte AJ, Ji HP, Snyder M. Nat Biotechnol. 2011;30(1):78–82. doi: 10.1038/nbt.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fisher RA. The genetical theory of natural selection. Oxford: Oxford University Press; 1930. [Google Scholar]
- 19.Katsonis P, Lichtarge O. Genome Research. 2014;24(12):2050–8. doi: 10.1101/gr.176214.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Illumina Platinum Genomes. 2015 http://www.illumina.com/platinumgenomes/
- 21.The 1000 Genomes Project Consortium. Nature. 2012;491:56–65. [Google Scholar]
- 22.Wang K, Li M, Hakonarson H. Nucleic Acids Research. 2010;38(16):e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orr HA. Nat Rev Genet. 2005;6(2):119–27. doi: 10.1038/nrg1523. [DOI] [PubMed] [Google Scholar]
- 24.Li H, Durbin R. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Racz C, Petrovski R, Saunders CT, Chorny I, Kruglyak S, Margulies EH, Chuang HY, Kallberg M, Kumar SA, Liao A, Little KM, Stromberg MP, Tanner SW. Bioinformatics. 2013;29(16):2041–3. doi: 10.1093/bioinformatics/btt314. [DOI] [PubMed] [Google Scholar]
- 26.Igbal Z, Caccamo M, Turner I, Flicek P, McVean G. Nat Genet. 2012;44(2):226–32. doi: 10.1038/ng.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DePristo M, Banks E, Poplin R, Garimella K, Maguire J, Hartl C, Philippakis A, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell T, Kernytsky A, Sivachenko A, Cibulskis K, Gabriel S, Altshuler D, Daly M. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]