Abstract
Purpose
New onset diabetes after transplantation (NODAT) is a serious complication following solid organ transplantation. There is a genetic contribution to NODAT and we have conducted comprehensive meta-analysis of available genetic data in kidney transplant populations.
Methods
Relevant articles investigating the association between genetic markers and NODAT were identified by searching PubMed, Web of Science and Google Scholar. SNPs described in a minimum of three studies were included for analysis using a random effects model. The association between identified variants and NODAT was calculated at the per-study level to generate overall significance values and effect sizes.
Results
Searching the literature returned 4,147 citations. Within the 36 eligible articles identified, 18 genetic variants from 12 genes were considered for analysis. Of these, three were significantly associated with NODAT by meta-analysis at the 5% level of significance; CDKAL1 rs10946398 p = 0.006 OR = 1.43, 95% CI = 1.11–1.85 (n = 696 individuals), KCNQ1 rs2237892 p = 0.007 OR = 1.43, 95% CI = 1.10–1.86 (n = 1,270 individuals), and TCF7L2 rs7903146 p = 0.01 OR = 1.41, 95% CI = 1.07–1.85 (n = 2,967 individuals).
Conclusion
Evaluating cumulative evidence for SNPs associated with NODAT in kidney transplant recipients has revealed three SNPs associated with NODAT. An adequately powered, dense genome-wide association study will provide more information using a carefully defined NODAT phenotype.
Introduction
New onset diabetes after transplantation (NODAT), also known as post transplantation diabetes mellitus (PTDM), is a serious complication of solid organ transplantation [1]. It affects 2–50%[1–3] of organ transplant recipients and is associated with greater healthcare costs and an increased risk of graft failure, cardiovascular complications and death [4]. The wide variation in reported prevalence of NODAT in part reflects the varying clinical definitions of this disorder. In different clinical studies the NODAT phenotype has been defined by various criteria including elevated fasting blood glucose; abnormal oral glucose tolerance tests; elevated glycated haemoglobin (HbA1c) or absolute requirement for hypoglycaemic therapies following solid organ transplantation [5,6]. A number of modifiable and non-modifiable risk factors have been identified which may predict NODAT. Modifiable risk factors include obesity and choice of anti-rejection immunosuppression medication [7]. Patients receiving tacrolimus-based immunosuppressive regimens are at greater risk of developing NODAT compared to those prescribed ciclosporin-based immunosuppressive treatment [8]. However, choosing an immunosuppressive regimen to specifically avoid NODAT may have a damaging effect on the graft itself [1]. Non-modifiable risk factors include family history of diabetes mellitus, polycystic kidney disease, hepatitis C infection, female gender and older recipient age [9,10]. There is an established genetic component to NODAT, however the identification of genetic risk factors has proved challenging. It is well documented that ethnicity is an important risk factor; people of African American, Hispanic, or South Asian background are at a significantly increased risk of developing the disease [5]. Low plasma adiponectin concentration, a factor which is under significant genetic control [11], has also been demonstrated to be predictive for NODAT [12]. Genome-wide association studies (GWAS) are revealing SNPs associated with diabetes, which are replicated across multiple populations [13,14], but such robust multi-centre GWAS have not yet been published for NODAT. However, multiple publications have reported genetic associations with NODAT in the literature, often with inconsistent results [1]; this report describes an inclusive review and meta-analysis of existing data.
Materials and Methods
Selection Criteria
Review of the literature was performed to identify all published genetic variants associated with NODAT in a kidney transplant population. Studies carried out in NODAT populations following other forms of organ transplant (such as liver transplant) were not included. PubMed, Web of Science and Google Scholar were searched from their inception until May 2015 with no language restrictions, using the following keywords: ‘New Onset Diabetes’, ‘Diabetes Mellitus’, ‘Gene’, ‘Genetic’, ‘Genotype’, ‘Transplantation’, ‘Transplant’, ‘Polymorphism’, ‘Mutation’, ‘NODAT’ and ‘PTDM’ (Post-Transplantation Diabetes Mellitus). Bibliographies for all identified articles and reviews were examined to identify further publications not found in the original search.
Inclusion Criteria
Studies were included when there was a minimum of three studies investigating the association of a specific variant with NODAT. Studies were deemed eligible if they fulfilled the following criteria: (a) published in a peer reviewed journal article or conference abstract using original data; (b) were conducted in a kidney transplant population in a case-control manner for NODAT; (c) included patients diagnosed with NODAT; (d) included controls who had undergone kidney transplantation but did not develop NODAT during follow-up. Authors were contacted if further essential information was required or if there was a query regarding eligibility. If sufficient information could not be obtained, the study was excluded, as were studies that duplicated data.
Statistical Analysis
Data was manually extracted from the studies. Information was gathered on study size, numbers of cases versus controls, ethnicity, genotyping methods, recorded odds ratios and p values. If ethnicity was not explicitly stated, this was inferred from the geographical location of the recruitment site and/or contact with authors. Deviation from Hardy-Weinberg Equilibrium was measured using genotype counts with a threshold of p <0.0001. Funnel plots of standard error of the log-odds-ratio against the log-odds-ratio were produced to estimate publication bias. Power calculations were conducted using StatCalc version 6.
Heterogeneity was calculated using a Cochrane Q test for heterogeneity with the I2 statistic used to describe percentage variation across studies. Meta-analysis was performed using a random effects model for variants replicated in three or more eligible studies, with significance value set at p <0.05.
All meta-analyses were performed using Review Manager software version 5.3.5 (RevMan 5.3) (http://tech.cochrane.org/revman)[15].
Funnel plots of standard error of the log-odds-ratio against the log-odds-ratio were produced to estimate publication bias. These were assessed by visual inspection. Funnel plots are capable of detecting publication bias which would be undetected by more formal statistical tests. Statistical tests such as Egger’s test were not conducted in this review due to the small number of studies in the meta-analyses, which were not sufficient to distinguish chance from asymmetry.
Results
Included Studies
The preliminary literature search yielded 4,147 citations, 40 of which were relevant studies investigating NODAT and 36 of which had all the required information to allow the extraction of variant information (Fig 1). Data was extracted from these articles for all investigated SNPs.
Fig 1. Flowchart describing the process of selection of eligible articles and variants for inclusion in the meta-analysis.

Of the 36 studies deemed eligible for inclusion, 16 studies were carried out in Asian populations, 16 in Caucasian populations and 4 in populations of mixed ethnicity. Table 1 outlines the characteristics of each of the eligible studies.
Table 1. Summary of eligible studies describing the ethnicity, genotyping method and total numbers of cases (NODAT patients) and controls (non-NODAT kidney transplant recipients).
| Paper | Ethnicity | Genotyping Method | NODAT Cases | Controls | Incidence | NODAT Definition |
|---|---|---|---|---|---|---|
| Babel 2004 [16] | Caucasian | PCR-SSP | 57 | 221 | 21% | Based on laboratory tests including FBG ≥126mg/dL |
| Cattaneo 2009 [17] | Caucasian | dHPLC & Direct Sequencing | 24 | 123 | 16% | PGC ≥200mg/dL with symptoms or FBG ≥126mg/dL |
| Chakkera 2012 [18] | Mixed | Sequenom iPLEX | 22 | 69 | 24% | Anti-diabetic medication requirement after one month post-transplant |
| Chang 2011 [19] | Asian | PCR-RFLP | 81 | 295 | 22% | FBG ≥126mg/dL or PGC ≥200mg/dL with symptoms or two hour PGC ≥200mg/dl during OGTT |
| Chen 2012 [20] | Asian | TaqMan & RT-PCR | 162 | 157 | 51% | Two or more FBG >126mg/dL or anti-diabetic medication requirement beyond 30 days post-transplant |
| Dutkiewicz 2010 [21] | Caucasian | PCR-RFLP | 21 | 138 | 13% | HbA1c continuously >6.5%, FBG ≥126mg/dL, or anti-diabetic medication requirement beyond three months post-transplant |
| Elens 2013 [22] | Caucasian | RT-PCR | 9 | 76 | 11% | Use of anti-diabetic medication at any time during the follow up period |
| Ergun 2011 [23] | Caucasian | PCR-RFLP | 9 | 73 | 11% | Symptoms of diabetes with PGC ≥200mg/dL, or two or more consecutive FBG >126mg/dL or two hour PGC ≥200mg/dL during OGTT |
| Fougeray 2011 [24] | Mixed | TaqMan | 14 | 255 | 5% | FBG >126mg/dL or non-fasting glycaemia >11mmol/L measured at baseline or at days 14, 30, 60, 90 |
| Ghisdal 2009 [25] | Caucasian | RT-PCR | 118 | 958 | 11% | FBG ≥126mg/dL on two or more occasions or de novo anti-diabetic medication requirement |
| Jeong 2010 [26] | Asian | Direct Sequencing (ABI-PRISM) | 56 | 255 | 18% | FPG >126g/dL, HbA1c >6.5 or insulin and oral hypoglycaemic agents required for over 3 months |
| Kang 2008 [27] | Asian | TaqMan | 174 | 450 | 28% | Three months post-transplant began anti-diabetic medication and continued thereafter |
| Kang 2008a [28] | Asian | TaqMan | 119 | 392 | 23% | One year post-transplant began anti-diabetic medication after continued thereafter |
| Kang 2009 [29] | Asian | RT-PCR | 145 | 444 | 25% | One year post-transplant began anti-diabetic medication after continued thereafter |
| Kang 2012 [30] | Asian | TaqMan | 154 | 421 | 27% | One year post-transplant began anti-diabetic medication after continued thereafter |
| Kao 2010 [31] | Asian | PCR-RFLP | 73 | 241 | 23% | Patients with HbA1c >6.5mg/dL on sequential blood samples |
| Khan 2015 [32] | Asian | PCR-RFLP | 42 | 98 | 30% | Administered anti-diabetic medication for more than three months post-transplant |
| Kim 2012 [33] | Asian | Direct Sequencing (ABI-PRISM) | 53 | 253 | 17% | FBG concentration over 125mg/dL, HbA1c more than 6.5% or anti-diabetic medication required for over 3 months |
| Kurzawski 2010 [34] | Caucasian | PCR method | 56 | 158 | 26% | HbA1c >6.5mg/dL, FBG >126mg/dL or those requiring anti-diabetic medication for greater than three months at one year post-transplant |
| Kurzawski 2011 [35] | Caucasian | RT-PCR | 66 | 168 | 28% | HbA1c >6.5mg/dL, FBG >126mg/dl or those requiring anti-diabetic medication for greater than three months |
| Kurzawski 2012 [36] | Caucasian | RT-PCR | 67 | 168 | 29% | HbA1c >6.5mg/dl, FBG >126mg/dL or those requiring anti-diabetic medication for greater than three months |
| Kurzawski 2014 [37] | Caucasian | RT-PCR | 48 | 176 | 21% | FPG >126mg/dL or those requiring anti-diabetic medication for greater than three months |
| Lee 2013 [38] | Asian | Direct Sequencing (ABI-PRISM) | 49 | 253 | 16% | Three months post-transplant FBG ≥126mg/dL or symptoms of diabetes with PGC ≥200mg/dL at any time of day or two hour PGC ≥200mg/dL during OGTT or anti-diabetic medication required for more than three months |
| McCaughan 2014 [1] | Caucasian | Illumina 660K Array & Sequenom iPLEX | 26 | 230 | 10% | New requirement for anti-diabetic medication after transplant |
| Nicoletto 2013 [39] | Caucasian | Sequenom iPLEX RT-PCR | 83 | 187 | 31% | Second recorded FBG of 126mg/dL or more |
| Özdemir 2011 [40] | Caucasian | PCR Method | 23 | 27 | 46% | Symptoms of diabetes with PGC ≥200mg/dL, or record of two or more consecutive FBG >126mg/dL or two hour PGC ≥200mg/dL during OGTT and anti-diabetic medication requirement |
| Szuszkiewicz 2011 [41] | Mixed | PCR-RFLP | 36 | 79 | 31% | Anti-diabetic medication requirement, FBG ≥126mg/dL and two hour PGC ≥200mg/dL when available from patient history |
| Tavira 2011 [42] | Caucasian | PCR-RFLP | 115 | 205 | 36% | FBG>126g/dL after three consecutive measurements |
| Tavira 2012 [43] | Caucasian | PCR-RFLP | 115 | 205 | 36% | FBG>126g/dL after three consecutive measurements |
| Tsai 2011 [44] | Asian | PCR-RFLP | 85 | 198 | 30% | FBG ≥126mg/dL or symptoms of diabetes and PGC ≥200mg/dL at any time of day or two hour PGC ≥200mg/dL during OGTT |
| Vattam 2013 [45] | Asian | PCR-RFLP | 42 | 98 | 30% | PGC ≥200mg/dL with diabetes symptoms or FBG ≥126mg/dL or two hour PGC ≥200mg/dL during OGTT |
| Wang 2011 [46] | Mixed | Direct Sequencing | 51 | 72 | 41% | FBG ≥126mg/dL three months post-transplant |
| Weng 2012 [47] | Asian | PCR-RFLP | 27 | 251 | 10% | PGC ≥200mg/dL with diabetes symptoms or FBG ≥126mg/dL or two hour PGC ≥200mg/dL during OGTT |
| Yang 2011 [48] | Caucasian | RT-PCR | 133 | 170 | 44% | Two or more occasions of FPG level >126mg/dL one month or more after transplant |
| Yao 2013 [49] | Asian | PCR-RFLP | 16 | 89 | 15% | PGC ≥200mg/dL with diabetes symptoms or FBG ≥126mg/dL or two hour PGC ≥200mg/dL during OGTT |
| Yu 2011 [50] | Asian | PCR-RFLP | 97 | 301 | 24% | FBG ≥126mg/dL on at least two occasions or to require anti-diabetic medication |
PCR-SSP, Polymerase chain reaction, single specific primer; dHPLC, Denaturing high performance liquid chromatography; RFLP, PCR Restriction Fragment Length Polymorphism; RT-PCR, Real Time PCR; FBG, Fasting Blood Glucose; PGC, Plasma Glucose Concentration; OGTT, Oral Glucose Tolerance Test; HbA1c, Haemoglobin A1c
The literature review revealed 18 genetic variants considered for association with NODAT that were reported in a minimum of three studies across 12 genes (Table 2 and Fig 2).
Table 2. Variants replicated in a minimum of three publications with associated odds ratios, 95% confidence intervals and p values following meta-analysis.
| Gene | Variant | Odds Ratio | 95% CI | p Value | Minor Allele | Minor Allele Frequency (Control Group) |
|---|---|---|---|---|---|---|
| CDKAL1 | rs10946398 | 1.43 | 1.11–1.85 | 0.006 | C | 42.65% |
| KCNQ1 | rs2237892 | 1.43 | 1.10–1.86 | 0.007 | T | 43.39% |
| TCF7L2 | rs7903146 | 1.41 | 1.07–1.85 | 0.01 | T | 18.41% |
| KCNJ11 | rs5219 | 1.28 | 0.92–1.76 | 0.14 | T | 35.50% |
| PPARG | rs4253728 | 1.55 | 0.78–3.11 | 0.21 | A | 23.71% |
| TNFA | rs1800629 | 0.81 | 0.56–1.17 | 0.25 | A | 14.07% |
| HHEX | rs1111875 | 1.14 | 0.89–1.44 | 0.30 | C | 49.92% |
| HHEX | rs5015480 | 1.24 | 0.77–1.97 | 0.38 | C | 33.70% |
| IGF2BP2 | rs1470579 | 1.15 | 0.84–1.59 | 0.39 | C | 33.26% |
| KCNJ11 | rs5215 | 1.09 | 0.88–1.34 | 0.42 | C | 36.07% |
| CDKN2A/B | rs10811661 | 1.1 | 0.79–1.54 | 0.57 | C | 25.87% |
| IGF2BP2 | rs4402960 | 0.96 | 0.80–1.14 | 0.61 | T | 31.85% |
| SLC30A8 | rs13266634 | 0.87 | 0.48–1.55 | 0.63 | T | 35.99% |
| PPARG | rs1801282 | 1.05 | 0.80–1.37 | 0.73 | G | 10.71% |
| TCF7L2 | rs12255372 | 1.06 | 0.77–1.47 | 0.73 | T | 24.11% |
| ADIPOQ | rs1501299 | 1.06 | 0.71–1.56 | 0.79 | T | 8.33% |
| CDKAL1 | rs7754840 | 1.04 | 0.79–1.37 | 0.80 | C | 31.50% |
| FTO | rs8050136 | 1.01 | 0.82–1.24 | 0.95 | A | 32.28% |
Variants highlighted in bold are those which reached significance at the 5% level.
Fig 2. Genetic variants explored for association with NODAT in at least three publications.

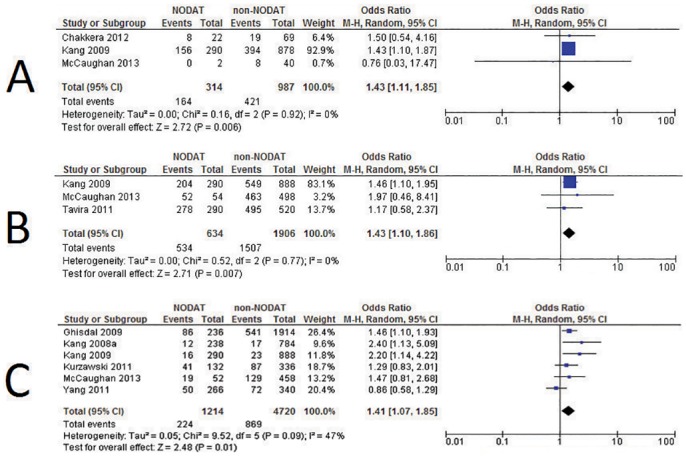
Of these analysed variants, three were significantly associated with NODAT based on meta-analysis; CDKAL1 rs10946398 p = 0.006 OR = 1.43, 95% CI = 1.11–1.85 (n = 696 individuals), KCNQ1 rs2237892 p = 0.007 OR = 1.43, 95% CI = 1.10–1.86 (n = 1,270 individuals), and TCF7L2 rs7903146 p = 0.01 OR = 1.41, 95% CI = 1.07–1.85 (n = 2,967 individuals) (Fig 3).
Fig 3. Forest Plots illustrating odds ratios and 95% confidence intervals for the three variants significantly associated with NODAT in random effects meta-analysis; black diamonds represent overall odds ratios for each of the variants. A CDKAL1 rs10946398 B KCNQ1 rs2237892 C TCF7L2 rs7903146.
Power calculations (S1 Table) suggest this study is adequately powered to identify a risk variant; for example, considering 2360 cases and 607 controls there was >80% power to identify a risk variant with odds ratio 1.5 and minor allele frequency of 5%.
Discussion
Main Findings
Thorough investigation of genetic variants reportedly associated with NODAT in kidney transplant patients has revealed significant associations by combined analysis. Three SNPs were significantly associated with NODAT–TCF7L2 rs7903146, CDKAL1 rs10946398 and KCNQ1 rs2237892 at the significance level p<0.05.
Many of the studies used in this investigation focused on genes previously associated with type 2 diabetes (T2D)[29,36]. NODAT and T2D have a number of important similarities. Both are characterised by insulin resistance and insulin hypo-secretion and share similar risk factors including increased age, family history of diabetes and non-white ethnicity [17].
TCF7L2 (transcription factor 7-like-2) has been previously linked to T2D, and has been cited as one of the most important signals associated with T2D [51]. The T allele was identified as a diabetes risk factor in the pre-GWAS era and was later replicated across a number of groups with different ethnic ancestry [52,53]. It is not yet completely understood how TCF7L2 influences risk of T2D but a number of theories have been put forward. It may affect blood glucose homeostasis by altering levels of glucagon-like peptide 1 in the gut, or it may decrease insulin secretion via the pancreatic beta, adipose or liver cells [54]. rs7903146 is located in an intron; a non-protein coding region of the gene [55]. There is no obvious mechanism by which a mutation at this locus could affect NODAT or T2D development, however the variant rs7903146 may either be in linkage disequilibrium with a causal allele or may itself influence gene expression through regulatory mechanisms.
CDKAL1 (cyclin dependent kinase 5 regulatory subunit associated protein 1 like 1) has been associated with impaired insulin secretion and the development of T2D in both European and Han Chinese populations by GWAS [56] and the variant rs10946398 has been found to be significantly associated with T2D by meta-analysis [57]. CDKAL1 encodes a methylthiotransferase which is thought to regulate the CDK5 protein which stimulates production of insulin as well as other processes in the pancreatic beta cells [58]. In this manner, by impairing insulin production via over-expression of CDK5, CDKAL1 may increase risk of T2D [57] and NODAT. The rs10946398 variant is found in exon 5 of the CDKAL1 gene. An alternative splicing product of CDKAL1 (CDKAL1v1) is increased in individuals homozygous for the minor C allele at this locus. It has therefore been suggested that this particular variant influences splicing of the gene [59].
KCNQ1 is also an established T2D risk factor and has been associated with gestational diabetes [60–62]. Variants of KCNQ1 cause a variety of disorders including hereditary long QT syndrome (Romano-Ward syndrome)[63]. It is expressed in the pancreatic islet cells as well as the heart and encodes a protein which combines with KCNE proteins to form voltage charged potassium channels found in the membranes. The KCNQ1 proteins form the structure of the channel while the KCNE proteins regulate the activity of the channel [64]. Pancreatic beta cell survival rate is thought to be affected by these potassium channels. It is thought that dysfunction of these potassium channels could alter cell membrane potential and contribute to development of T2D or NODAT. A specific KCNQ1 blocker 293B has been shown to increase insulin production [65]. The variant rs2237892 C risk allele has been shown to be associated with fasting plasma glucose concentration, suggesting that C homozygous individuals have impaired baseline insulin secretion. The gene is also under the control of tissue specific imprinting [66].
These genetic variants are all established T2D risk factors and several variants have been implicated in potential mechanisms contributing to diabetes. Therefore, it is not surprising that these variants are linked to NODAT, another form of diabetes, since the mechanisms controlling insulin production and maintenance of stable glucose levels will both be similar in T2D and NODAT. The meta-analyses conducted on the other variants identified in the literature did not reach statistical significance. This may have been for several reasons, including the small number of studies, small numbers of study participants, or varying phenotypic definitions. Of note, our meta-analysis incorporates data from both candidate gene and genome-wide association studies.
A number of variants which were associated with NODAT in previous studies were not found to be associated with NODAT following meta-analysis. Notable variants which did not reach significance after meta-analysis include KCNJ11 rs5219 and ADIPOQ rs1501299. KCNJ11 rs5219 is an established T2D risk factor in a gene encoding a voltage gated potassium channel. ADIPOQ rs1501299 has been previously associated with NODAT as well as breast cancer, prostate cancer and T2D complications such as heart disease. Adiponectin encoded by ADIPOQ is involved in lipid metabolism and insulin sensitivity and making this an attractive candidate for association with NODAT. Neither of these particular variants reached the p<0.05 significance threshold following meta-analysis which could mean they are not associated with NODAT or that the association is only present in certain populations.
Limitations of the Review
This study does have a number of limitations. The definition of NODAT differs from centre to centre as highlighted by the varying prevalence of NODAT reported in Table 1 which ranges from 5–51%. A potential explanation for this large variation in reported prevalence is the differences in how NODAT is diagnosed i.e. heterogeneity of the clinical phenotype. Some authors employed diagnostic criteria for diabetes as defined by the World Health Organisation and American Diabetic Association, although the final interpretation of these standards did vary in published studies of NODAT. Others authors used pragmatic clinical criteria for NODAT diagnosis defining the affected patients as requiring the de novo prolonged use of insulin and/or oral hypoglycaemic medication following transplant. A more rigorously defined NODAT phenotype may facilitate more reproducible results between studies. There were a small number of studies available for many of the variants; larger, carefully phenotyped studies would provide better power to identify alleles robustly associated with NODAT. There was a varying degree of heterogeneity noted between studies, some of which was likely due to different ethnicities considered. In addition, the variations in prescribed immunosuppressive regimens, and their differential effects on NODAT incidence, were not accounted for in many of the studies. It is of note that the Belfast derived data was from the single transplant centre for Northern Ireland. TCF7L2 rs7903146 variant was only nominally associated with NODAT (p = 0.01), but this association was replicated with the same direction of effect across five independent collections. Possible interactions among the genetic variants identified have not been investigated and this is a further limitation of the study.
Conclusions
This is a thorough overview of all reported genetic factors influencing the development of NODAT in the current literature. Analysis revealed a significant association between NODAT and three established T2D risk factor variants. Functional studies will be required to further investigate these variants and associated pathways to gain a complete perspective on their effects. In order to obtain more consistency between studies and identify risk alleles with smaller effect sizes, larger participant numbers through multi-centre collaboration and harmonised phenotypic definition of NODAT is required. An adequately powered, dense genome-wide association study will provide more information using a carefully defined NODAT phenotype.
Hypothesis free approaches such as the GWAS carried out by McCaughan and colleagues for NODAT are advantageous to identify new biological targets and therapeutic pathways and should also be carried out in other populations and ethnicities to better understand the genetic architecture underlying this disease [1].
Supporting Information
This table describes the power which this study had to identify significant genetic variants. The power was based on 607 cases, 2360 controls and dependent on the Minor Allele Frequency, significance sought, and effect size (reported for each variant in Table 2).
(XLSX)
Describes how this manuscript conforms to the PRISMA 2009 guidelines.
(DOCX)
Acknowledgments
KAB is supported by a Queen’s University Belfast, School of Medicine, Dentistry and Biomedical Science International PhD Fellowship as well as receiving support from the Northern Ireland Kidney Research Fund. The NIKRF had no role in the study design.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
KAB is supported by a Queen's University Belfast, School of Medicine Dentistry and Biomedical Sciences (http://www.qub.ac.uk/schools/mdbs/) international studentship and the Northern Ireland Kidney Research Fund, a local charity (http://www.kidneyresearchni.com/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.McCaughan J, McKnight AJ, Maxwell AP. Genetics of new-onset diabetes after transplantation. J Am Soc Nephrol. 2014;25(5):1037–49. 10.1681/ASN.2013040383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mora PF. New-onset diabetes after renal transplantation. J Investig Med. 2010;58(6):755–63. [DOI] [PubMed] [Google Scholar]
- 3.Kesiraju S, Paritala P, Rao Ch UM, Sahariah S. New onset of diabetes after transplantation—An overview of epidemiology, mechanism of development and diagnosis. Transpl Immunol. 2014;30(1):52–8. [DOI] [PubMed] [Google Scholar]
- 4.Hecking M, Werzowa J, Haidinger M, Hörl WH, Pascual J, Budde K, et al. Novel views on new-onset diabetes after transplantation: Development, prevention and treatment. Nephrol Dial Transplant. 2013;28(3):550–66. 10.1093/ndt/gfs583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davidson J., Wilkinson A. New-onset diabetes after transplantation 2003 international consensus guidelines: An endocrinologist’s view. Diabetes Care. 2004;27(3):805–12. [DOI] [PubMed] [Google Scholar]
- 6.Shabir S, Jham S, Harper L, Ball S, Borrows R, Sharif A. Validity of glycated haemoglobin to diagnose new onset diabetes after transplantation. Transpl Int. 2013;26(3):315–21 10.1111/tri.12042 [DOI] [PubMed] [Google Scholar]
- 7.Sharif A, Hecking M, De Vries AP, Porrini E, Hornum M, Rasoul-Rockenschaub S, et al. Proceedings from an international consensus meeting on post transplantation diabetes mellitus: Recommendations and future directions. Am J Transplant. 2015;14(9):1992–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luan FL, Steffick DE, Ojo AO. New-onset diabetes mellitus in kidney transplant recipients discharged on steroid-free immunosuppression. Transplantation. 2011:91(3):334–41 [DOI] [PubMed] [Google Scholar]
- 9.Gaynor JJ, Ciancio G, Guerra G, Sageshima J, Hanson L, Roth D, et al. Multivariable risk of developing new onset diabetes after transplant-results from a single-center study of 481 adult, primary kidney transplant recipients. Clin Transplant. 2015;29(4):301–10. 10.1111/ctr.12510 [DOI] [PubMed] [Google Scholar]
- 10.Palepu S. New-onset diabetes mellitus after kidney transplantation: Current status and future directions. World J Diabetes. 2015;6(3):445 10.4239/wjd.v6.i3.445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menzaghi C, Ercolino T, Salvemini L, Coco A, Kim SH, Fini G, et al. Multigenic control of serum adiponectin levels: Evidence for a role of the APM1 gene and a locus on 14q13. Physiol Genomics. 2004;19(2):170–4. [DOI] [PubMed] [Google Scholar]
- 12.Wiecek A. Genotypes of renin-angiotensin system and plasma adiponectin concentration in kidney transplant patients. Ann Transplant. 2013;18:593–603. 10.12659/AOT.884022 [DOI] [PubMed] [Google Scholar]
- 13.Morris A, Voight B, Teslovich T. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44(9):981–90. 10.1038/ng.2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hara K, Fujita H, Johnson T, Yamauchi T, Yasuda K, Horikoshi M, et al. Genome-wide association study identifies three novel loci for type 2 diabetes. Hum Mol Genet. 2014;23(1):239–46. 10.1093/hmg/ddt399 [DOI] [PubMed] [Google Scholar]
- 15.The Cochrane Collaboration. Review Manager (RevMan) [Computer program]. Version 5.3. 2014.
- 16.Babel N, Cherepnev G, Kowalenko A, Horstrup J, Volk H-D, Reinke P. Nonimmunologic complications and gene polymorphisms of immunoregulatory cytokines in long-term renal transplants. Kidney Int. 2004;66:428–32. [DOI] [PubMed] [Google Scholar]
- 17.Cattaneo D, Ruggenenti P, Baldelli S, Motterlini N, Gotti E, Sandrini S, et al. ABCB1 genotypes predict cyclosporine-related adverse events and kidney allograft outcome. J Am Soc Nephrol. 2009;20(6):1404–15. 10.1681/ASN.2008080819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chakkera HA, Hanson RL, Raza SM, Distefano JK, Millis MP, Heilman RL, et al. Pilot Study: Association of traditional and genetic risk factors and new-onset diabetes mellitus following kidney transplantation. Transplant Proc. 2012;41(10):4172–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang HR, Yang SF, Tsai JP, Hsieh MC, Wu SW, Tsai HC, et al. Plasminogen activator inhibitor-1 5G/5G genotype is a protecting factor preventing post transplant diabetes mellitus. Clin Chim Acta. 2011;412(3–4):322–6. 10.1016/j.cca.2010.10.029 [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Sampaio M, Yang J, Min D, Hutchinson I. Genetic polymorphisms of the transcription factor NFATc4 and development of new-onset diabetes after transplantation in Hispanic kidney transplant recipients. Transplantation. 2012;93(3):325–30. [DOI] [PubMed] [Google Scholar]
- 21.Dutkiewicz G, Domanski L, Pawlik A, Binczak-Kuleta A, Safranow K, Ciechanowicz A, et al. Polymorphisms of superoxide dismutase, glutathione peroxidase and catalase genes in patients with post-transplant diabetes mellitus. Arch Med Res. 2010;41(5):350–5. 10.1016/j.arcmed.2010.06.006 [DOI] [PubMed] [Google Scholar]
- 22.Elens L, Sombogaard F, Hesselink D, van Schaik RHN, van Gelder T. Single-nucleotide polymorphisms in P450 oxidoreductase and peroxisome proliferator-activated receptor-α are associated with the development of new-onset diabetes after transplantation in kidney transplant recipients treated with tacrolimus. Pharmacogenet Genomics. 2013;23(12):649–57. [DOI] [PubMed] [Google Scholar]
- 23.Ergün I, Keven K, Sengül S, Karabulut HG, Kurultak I, Soypacaci Z, et al. Endothelial nitric oxide synthase gene intron 4 polymorphism predicts new onset diabetes mellitus after transplantation in kidney allograft recipients treated with cyclosporin A. Int Urol Nephrol. 2011;43(2):543–8. 10.1007/s11255-010-9786-8 [DOI] [PubMed] [Google Scholar]
- 24.Fougeray S, Loriot M, Nicaud V, Legendre C, Thervet E, Pallet N. Increased body mass index after kidney transplantation in activating transcription factor 6 single polymorphism gene carriers. Transplant Proc. 2011;43(9):3418–22. 10.1016/j.transproceed.2011.09.022 [DOI] [PubMed] [Google Scholar]
- 25.Ghisdal L, Baron C, Le Meur Y, Lionet A, Halimi JM, Rerolle JP, et al. TCF7L2 polymorphism associates with new-onset diabetes after transplantation. J Am Soc Nephrol. 2009;20(11):2459–67. 10.1681/ASN.2008121314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeong KH, Moon JY, Chung JH, Kim YH, Lee TW. Significant Associations between CCL5 polymorphisms and post transplantation diabetes mellitus in Korean renal allograft recipients. Am J Nephrol. 2010;32:356–61. 10.1159/000319704 [DOI] [PubMed] [Google Scholar]
- 27.Kang ES, Nam CM, Kim MS, Ahn CW, Kim YS, Cha BS, et al. A variant of the transcription factor 7- Like 2 (TCF7L2) gene and the risk of post transplantation diabetes mellitus in renal allograft recipients. Diabetes Care. 2008;31(1):63–8. [DOI] [PubMed] [Google Scholar]
- 28.Kang ES, Kim SM, Kim YS, Kim CH, Han SJ, Chun SW, et al. A polymorphism in the zinc transporter gene SLC30A8 confers resistance against post transplantation diabetes mellitus in renal allograft recipients. Diabetes. 2008;57(4):1043–7. [DOI] [PubMed] [Google Scholar]
- 29.Kang ES, Kim SM, Kim CH, Chung MN, Han SJ, Hur KY, et al. Association of common type 2 diabetes risk gene variants and post transplantation diabetes mellitus in renal allograft recipients in Korea. Transplantation. 2009;88(5):693–8. [DOI] [PubMed] [Google Scholar]
- 30.Kang ES, Magkos F, Kim BS, Zhai R, Su L, Kim YS, et al. Variants of the adiponectin and adiponectin receptor-1 genes and post transplantation diabetes mellitus in renal allograft recipients. J Clin Endocrinol Metab. 2012;97(1):E129–35. 10.1210/jc.2011-1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kao CC, Lian JD, Chou MC, Chang HR, Yang SF. Tumor necrosis factor alpha promoter polymorphism in post transplantation diabetes mellitus of renal transplant recipients. Transplant Proc. 2010;42(9):3559–61. 10.1016/j.transproceed.2010.06.032 [DOI] [PubMed] [Google Scholar]
- 32.Khan IA, Vattam KK, Jahan P, Mukkavali KK, Hasan Q, Rao P. Correlation between KCNQ1 and KCNJ11 gene polymorphisms and type 2 and post transplant diabetes mellitus in the Asian Indian population. Genes & Diseases; 2015;(Corrected Proof):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim YG, Ihm CG, Lee TW, Lee SH, Jeong KH, Moon JY, et al. Association of genetic polymorphisms of interleukins with new-onset diabetes after transplantation in renal transplantation. Transplantation. 2012;93(9):900–7. [DOI] [PubMed] [Google Scholar]
- 34.Kurzawski M, Dziewanowski K, Kedzierska K, Gornik W, Banas A, Drozdzik M. Association of calpain-10 gene polymorphism and post transplant diabetes mellitus in kidney transplant patients medicated with tacrolimus. Pharmacogenomics J. 2010;10(2):120–5. 10.1038/tpj.2009.44 [DOI] [PubMed] [Google Scholar]
- 35.Kurzawski M, Dziewanowski K, Kêdzierska K, Wajda A, Lapczuk J, Droÿdzik M. Association of transcription factor 7-like 2 (TCF7L2) gene polymorphism with post transplant diabetes mellitus in kidney transplant patients medicated with tacrolimus. Pharmacol Reports. 2011;63:826–33. [DOI] [PubMed] [Google Scholar]
- 36.Kurzawski M, Dziewanowski K, Łapczuk J, Wajda A, Droździk M. Analysis of common type 2 diabetes mellitus genetic risk factors in new-onset diabetes after transplantation in kidney transplant patients medicated with tacrolimus. Eur J Clin Pharmacol. 2012;68(12):1587–94. 10.1007/s00228-012-1292-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurzawski M, Malinowski D, Dziewanowski K, Droździk M. Impact of PPARA and POR polymorphisms on tacrolimus pharmacokinetics and new-onset diabetes in kidney transplant recipients. Pharmacogenet Genomics. 2014;24:397–400. [DOI] [PubMed] [Google Scholar]
- 38.Lee SR, Moon JY, Lee SH, Ihm CG, Lee TW, Kim SK, et al. Angiotensinogen polymorphisms and post-transplantation diabetes mellitus in Korean renal transplant subjects. Kidney Blood Press Res. 2013;37(2–3):95–102. 10.1159/000343404 [DOI] [PubMed] [Google Scholar]
- 39.Nicoletto BB, Souza GC, Fonseca NKO, Centenaro A, Manfro RC, Canani LHS, et al. Association between 276G/T adiponectin gene polymorphism and new-onset diabetes after kidney transplantation. Transplantation. 2013;96(12):1059–64. [DOI] [PubMed] [Google Scholar]
- 40.Özdemir BH, Özdemir FN, Ataç FB, Özdemir AA, Haberal M. Angiotensinogen t235 and angiotensin-converting enzyme insertion/deletion polymorphisms associated with the development of post transplantation diabetes mellitus in renal allograft recipients. Transplant Proc. 2011;43(2):572–4. 10.1016/j.transproceed.2011.01.046 [DOI] [PubMed] [Google Scholar]
- 41.Szuszkiewicz M, Bell J, Vazquez M, Adams-Huet B, Grundy SM, Chandalia M, et al. ENPP1/PC-1 K121Q and other predictors of post transplant diabetes. Metab Syndr Relat Disord. 2011; 9(1):25–9. 10.1089/met.2010.0041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tavira B, Diaz-Corte C, Ortega F, Arias M, Torres A, Diaz J, et al. KCNQ1 gene variants and risk of new-onset diabetes in tacrolimus-treated renal-transplanted patients. Clin Transplant. 2011;25:E284–91. 10.1111/j.1399-0012.2011.01417.x [DOI] [PubMed] [Google Scholar]
- 43.Tavira B, Coto E, Torres A, Díaz-Corte C, Díaz-Molina B, Ortega F, et al. Association between a common KCNJ11 polymorphism (rs5219) and new-onset post transplant diabetes in patients treated with Tacrolimus. Mol Genet Metab. 2012;105(3):525–7. 10.1016/j.ymgme.2011.12.020 [DOI] [PubMed] [Google Scholar]
- 44.Tsai JP, Yang SF, Wu SW, Hung TW, Tsai HC, Lian JD, et al. Glutathione S-transferase gene polymorphisms are not major risks for susceptibility to post transplantation diabetes mellitus in Taiwan renal transplant recipients. J Clin Lab Anal. 2011;25(6):432–5. 10.1002/jcla.20498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vattam KK, Khan IA, Movva S, Mukkavali KK, Poornima S, Rao P, et al. IGF2 ApaI A/G polymorphism evaluated in ESRD individuals as a biomarker to identify patients with new onset diabetes mellitus after renal transplant in Asian Indians. Open J Nephrol. 2013; 3, 104 [Google Scholar]
- 46.Wang P, Hudspeth E. Increased body mass index but not common vitamin D receptor, peroxisome proliferator-activated receptor γ, or cytokine polymorphisms confers predisposition to post transplant diabetes. Arch Pathol Lab Med. 2011;135(12):1581–4. 10.5858/arpa.2011-0160-OA [DOI] [PubMed] [Google Scholar]
- 47.Weng SC, Shu KH, Tarng DC, Wu MJ, Chen CH, Yu TM, et al. Gene polymorphisms are associated with post transplantation diabetes mellitus among Taiwanese renal transplant recipients. Transplant Proc. 2012;44(3):667–71. 10.1016/j.transproceed.2011.11.011 [DOI] [PubMed] [Google Scholar]
- 48.Yang J, Hutchinson II, Shah T, Min DI. Genetic and clinical risk factors of new-onset diabetes after transplantation in Hispanic kidney transplant recipients. Transplantation. 2011;91:1114–9. [DOI] [PubMed] [Google Scholar]
- 49.Yao B, Chen X, Shen FX, Xu W, Dong TT, Chen LZ, et al. The incidence of post transplantation diabetes mellitus during follow-up in kidney transplant recipients and relationship to Fok1 vitamin D receptor polymorphism. Transplant Proc. 2013;45(1):194–6. 10.1016/j.transproceed.2012.08.019 [DOI] [PubMed] [Google Scholar]
- 50.Yu AR, Xin HW, Wu XC, Fan X, Liu HM, Li G, et al. Adiponectin gene polymorphisms are associated with post transplantation diabetes mellitus in Chinese renal allograft recipients. Transplant Proc. 2011;43(5):1607–11. 10.1016/j.transproceed.2011.02.057 [DOI] [PubMed] [Google Scholar]
- 51.Hattersley AT. Prime suspect : the TCF7L2 gene and type 2 diabetes risk. Nephrology. 2007;117(8):6–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38(3):320–3. [DOI] [PubMed] [Google Scholar]
- 53.Tong Y, Lin Y, Zhang Y, Yang J, Zhang Y, Liu H, et al. Association between TCF7L2 gene polymorphisms and susceptibility to type 2 diabetes mellitus: a large Human Genome Epidemiology (HuGE) review and meta-analysis. BMC Med Genet. 2009;10:15 10.1186/1471-2350-10-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dormans JP, Gunawardena AT, Hakonarson H, Hecht JT. The type 2 diabetes associated rs7903146 T allele within TCF7L2 is significantly under-represented in hereditary multiple exostoses: insights into pathogenesis. Bone. 2015;(72):123–7. 10.1016/j.bone.2014.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nicod N, Pradas-Juni M, Gomis R. Role of the single nucleotide polymorphism rs7903146 of TCF7L2 in inducing nonsense-mediated decay. Springerplus. 2014: January 22;3:41 10.1186/2193-1801-3-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steinthorsdottir V, Thorleifsson G, Reynisdottir I, Benediktsson R, Jonsdottir T, Walters GB, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet. 2007;39(6):770–5. [DOI] [PubMed] [Google Scholar]
- 57.Dehwah MS, Wang M, Huang QY. CDKAL1 and type 2 diabetes: A global meta-analysis. Genet Mol Res. 2010;9(2):1109–20. 10.4238/vol9-2gmr802 [DOI] [PubMed] [Google Scholar]
- 58.Brambillasca S, Altkrueger A, Colombo SF, Friederich A, Eickelmann P, Mark M, et al. CDK5 regulatory subunit-associated protein 1-like 1 (CDKAL1) is a tail-anchored protein in the endoplasmic reticulum (ER) of insulinoma cells. J Biol Chem. 2012;287(50):41808–19 10.1074/jbc.M112.376558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou B, Wei FY, Kanai N, Fujimura A, Kaitsuka T, Tomizawa K. Identification of a splicing variant that regulates type 2 diabetes risk factor CDKAL1 level by a coding-independent mechanism in human. Hum Mol Genet. 2014;23(17):4639–50 10.1093/hmg/ddu184 [DOI] [PubMed] [Google Scholar]
- 60.Bazzi MD, Nasr FA, Alanazi MS, Alamri A, Turjoman AA. Association between FTO, MCR, SLC30A8, and KCNQ1 gene variants and type 2 diabetes in Saudi population. Genet Mol Res. 2014;1(4):10194–203. [DOI] [PubMed] [Google Scholar]
- 61.Ao D, Wang H, Wang L, Song J, Yang H, Wang Y. The rs2237892 Polymorphism in KCNQ1 influences gestational diabetes mellitus and glucose levels: A case-control study and meta-analysis. PLoS One. 2015;10(6):e0128901 10.1371/journal.pone.0128901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huerta-Chagoya A, Vázquez-Cárdenas P, Moreno-Macías H. Genetic determinants for gestational diabetes mellitus and related metabolic traits in Mexican women. PLoS One. 2015;10(5):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tester DJ, Ackerman MJ. Genetics of long Qt syndrome. Methodist Debakey Cardiovasc J. 2014;10(1):29–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abbot GW. KCNE1 and KCNE3: The yin and yang of voltage-gated K+ channel regulation. Gene;2015:S0378-119(15)01168-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lui L, Wang F, Lu H, Ren X, Zou J. Chromanol 293B, an inhibitor of KCNQ1 channels, enhances glucose-stimulated insulin secretion and increases glucagon-like peptide-1 level in mice. Islets. 2014;6(4) e962386 10.4161/19382014.2014.962386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schultz B, Gallicio G, Cesaroni M, Lupey L, Engel N. Enhancers compete with a long non-coding RNA for regulation of the Kcnq1 domain. Nucl. Acids Res. 2015; 43 (2): 745–759 10.1093/nar/gku1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This table describes the power which this study had to identify significant genetic variants. The power was based on 607 cases, 2360 controls and dependent on the Minor Allele Frequency, significance sought, and effect size (reported for each variant in Table 2).
(XLSX)
Describes how this manuscript conforms to the PRISMA 2009 guidelines.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.


