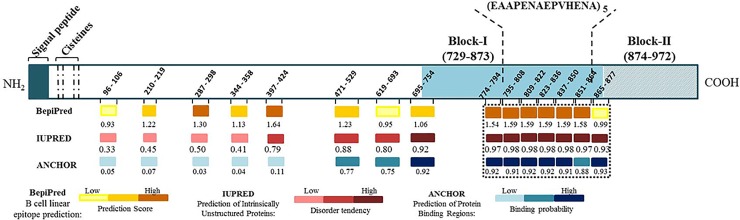
Fig 1. Schematic diagram of PvMSP9 and the predictions scores for linear B cell epitopes, intrinsically unstructured/disordered regions and protein-protein interaction regions.
The region corresponding to the amino acid residues 795–808 of PvMSP9 was selected for the synthesis of a soluble peptide based on the best combination of prediction scores using BepiPred, IUPRED and ANCHOR algorithms. Yellow heat bars represent B-cell epitopes, red heat bars represent predicted unordered regions and blue heat bars represent prediction of binding regions. The prediction scores represents the average of scores for all amino acids within the region with prediction values above the cut-offs chosen for significance. The bar color intensities are proportional to the prediction scores.