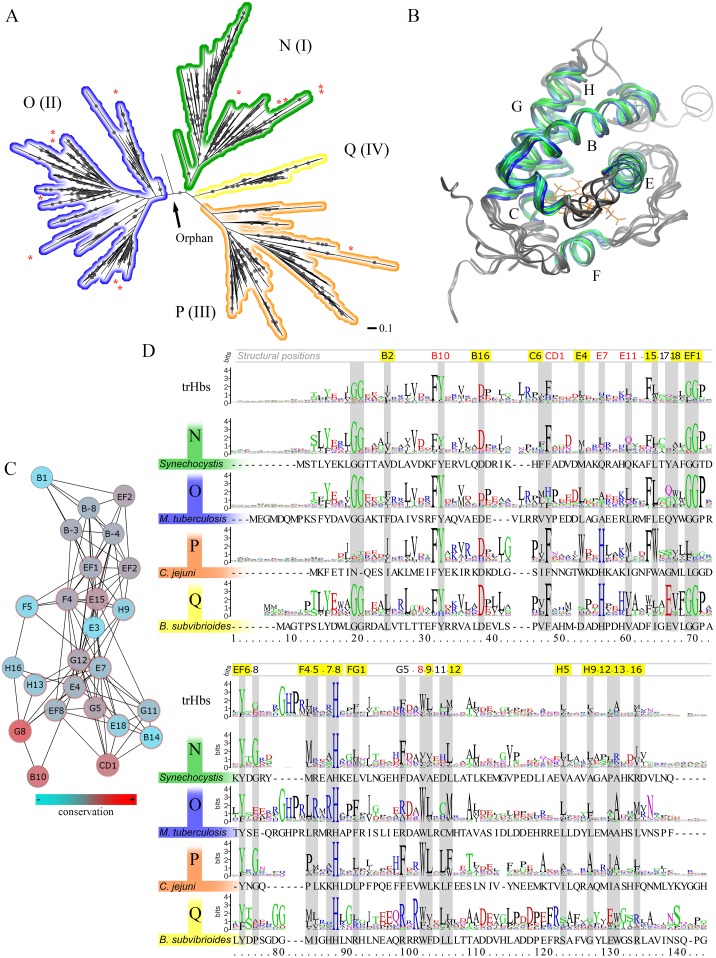
Fig 2. Phylogenetic hallmarks of trHbs sequences.
(A) Bayesian phylogenetic tree of trHbs family presented as a radial phylogram. Dots indicate bayesian support of ≥80%. The scale bar represents a distance of 0.1 accepted amino acid substitutions per site. Red asterisks indicate sequences with resolved 3D structures that were used in the construction of both the multiple sequence alignment (MSA) used for phylogeny and the structural alignment presented in (B). (B) Structural alignment of 17 available trHbs sequences. The selected color regions, together with helix labeling, correspond with the most conserved regions in the MSA. (C) MI network of positions that have cMI > 65. Specificity determining position (SDP) E7 and its direct neighbors are highlighted. Color of the node is indicative for the Kullback-Leibler information. Red encircled nodes correspond with the central SDP, E7, and its direct neighbors. (D) Logo sequences trimmed with block mapping and gathering with entropy (BMGE) for all 1107 considered trHbs and for each separate group. One sequence member of each subgroup is also shown as a reference. Known hallmark and other important positions are shaded in grey with their structural position denominators indicated above, in red for active site residues and with yellow shading for identified SDPs.