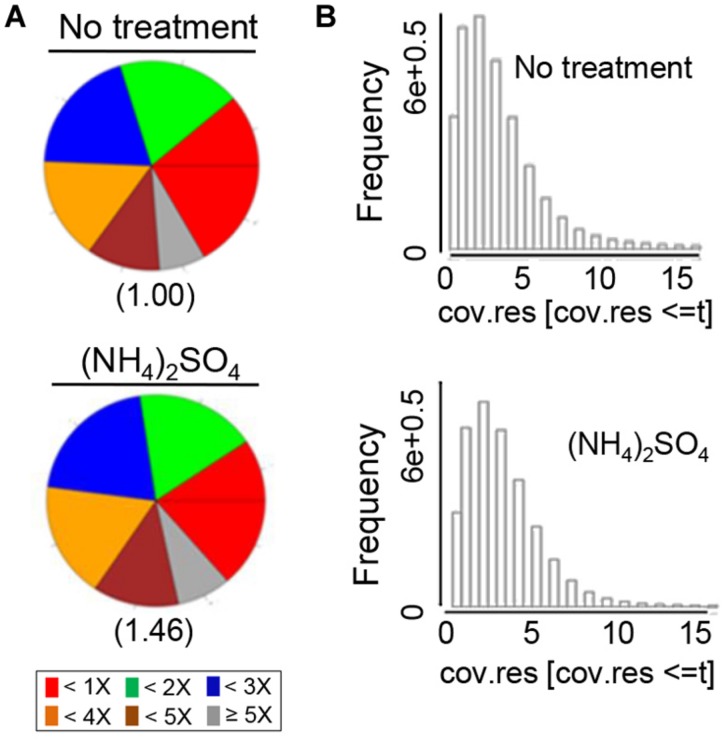
FIGURE 1.
MeDIP sequencing was used to detect whole genome methylation in ammonium-treated and untreated control Arabidopsis plants. Genomic coverage was quantified by the number of DNA methylation measurements that overlapped with CpG islands. (A) CpG coverage was determined by counting the number of reads. CpG islands were identified using the program MEDIPS. The numbers below the diagrams indicate “pattern not covered.” (B) The methylation pattern is presented as sequence pattern coverage. The parameter t indicates the maximal coverage depth to be plotted (15 reads per sequence pattern).