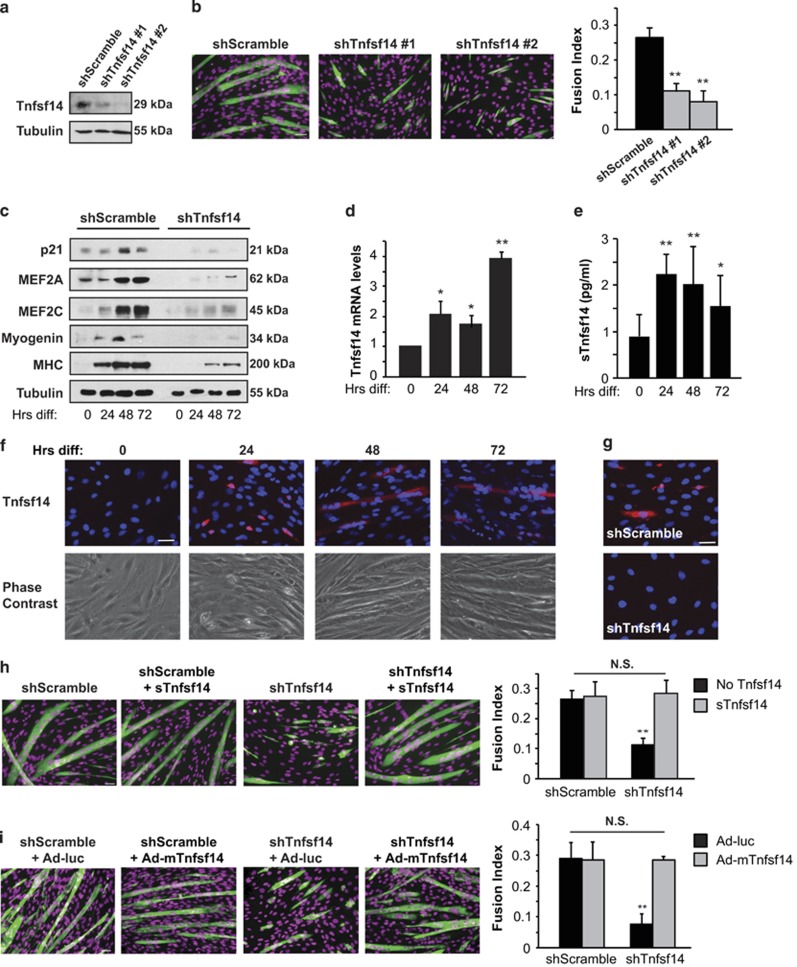
Figure 1.
Tnfsf14 is a positive regulator of myoblast differentiation. (a) C2C12 cells were infected with lentiviruses expressing shTnfsf14 or shScramble (negative control), selected for 2 days, followed by cell lysis and western analysis (n=4). (b) Cells treated as in (a) were differentiated for 72 h, followed by staining for MHC (green) and DAPI (pseudo-colored magenta), and quantification of fusion index (n=5). (c) Cells treated as in (a) were differentiated, and at indicated time points ('Hrs diff') were lysed and subjected to western analysis (n=4). (d) RNA was isolated from differentiating C2C12 cells at indicated time points and subjected to qRT-PCR analysis for Tnfsf14 mRNA levels (n=6). (e) Cell media over the course of differentiation were subjected to ELISA assay to determine sTnfsf14 levels (n=8). (f) Differentiating C2C12 cells at various time points ('Hrs diff') were stained without permeabilization for Tnfsf14 (red) and DAPI (blue) (n=4). (g) Cells treated as in a were differentiated for 24 h, followed by staining for Tnfsf14 (red) and DAPI (blue) (n=3). (h) C2C12 cells were treated as in (a), and then differentiated in the presence or absence of 25 ng/ml recombinant sTnfsf14 for 3 days, followed by staining for MHC and DAPI, and quantification of fusion index (n=3). (i) C2C12 cells were treated as in (a), and then infected with adenoviruses expressing mTnfsf14 or luciferease (luc; negative control), followed by differentiation for 3 days and then staining for MHC and DAPI. The fusion index was quantified (n=3). All error bars represent S.D. of independent replicates. One-sample two-tailed t-test was performed for data in (d), and paired two-tailed t-tests for all other data. *P<0.05; **P<0.01. Scale bars: 50 μm