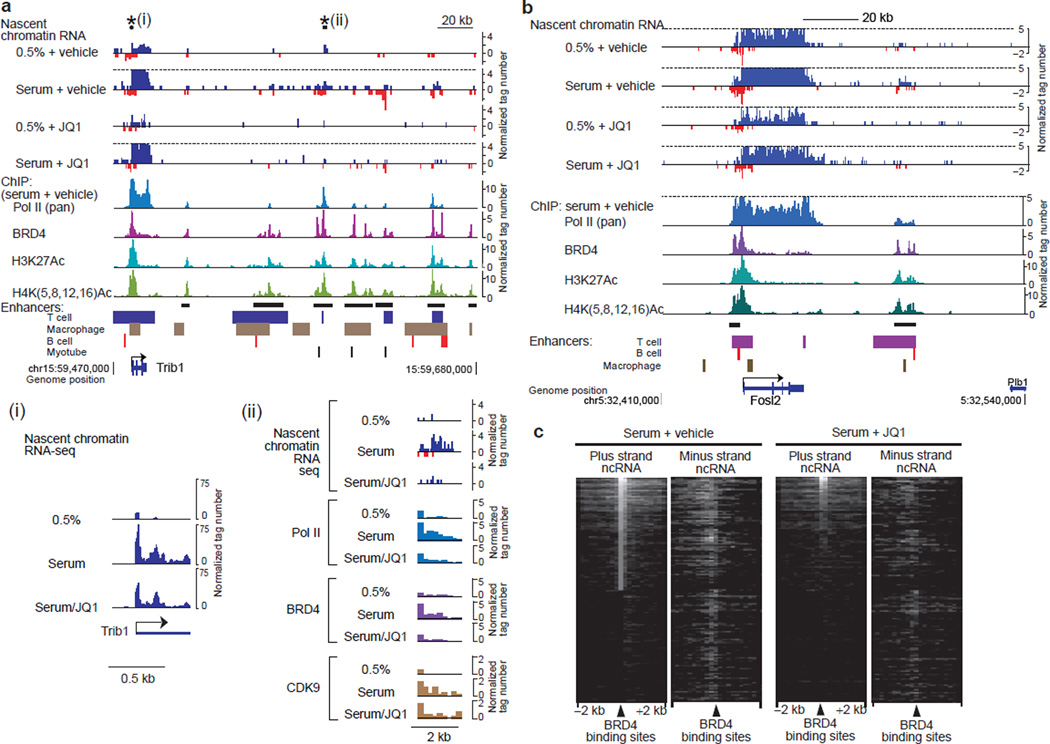
Figure 2. BET inhibitor JQ1 antagonized BRD4 and inhibits eRNA synthesis.
a, Genome browser views of chromatin-bound RNA-seq reads along the Trib1 gene and its downstream intergenic region. Sense- and antisense-strand signals are shown in upward and downward directions, respectively. Signals higher than the dotted lines are cut. ChIP-seq reads of Pol II, BRD4, H3K27Ac and H4Ac in serum+vehicle cells and known enhancer regions42 are aligned below, and potential enhancer clusters are marked by horizontal black bars. The regions marked with asterisks are shown expanded in insets (i) and (ii). Inset (i) shows rescaled views of nascent RNA-seq reads around the TSS. Inset (ii) shows expanded views of nascent RNA-seq reads and ChIP-seq reads of Pol II, BRD4 and CDK9. b, Genome browser views along the Fosl2 gene and the flanking regions. c, Heat map of chromatin-bound non-coding RNA-seq reads around intergenic BRD4 binding sites. ncRNA: non-coding RNA.