Fig. 4.
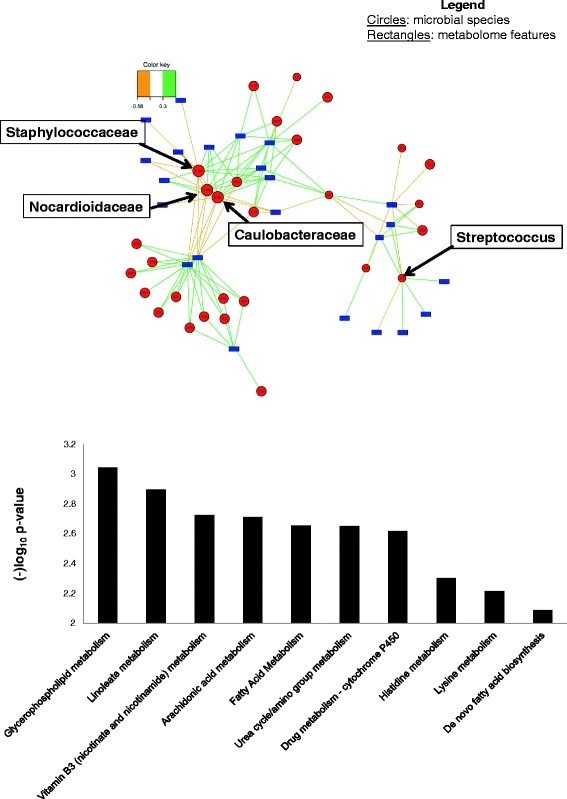
a Three-way relationship between metabolome, HIV, and microbiota integration of HIV-specific m/z features with microbiota data at correlation of 0.3. This figure shows that of the 91 m/z features associated with HIV, 23 were also found to be associated with 29 microbial genera at a correlation of 0.30; red circles microbial families and genus, blue rectangles metabolome features. b Pathway analysis using m/z features that were associated with Staphylococcaceae in the global network. Using Mummichog, metabolite annotation and pathway enrichment analysis was done using the m/z features that were associated with Staphylococcaceae (as the representative bacteria since pathways for Caulobacteraceae and Nocardioidaceae were very similar). The y-axis represents the enrichment significance of the metabolic pathways. This analysis shows that the pathways most affected included lineolate, glycerophospholipid, and fatty acid metabolism
