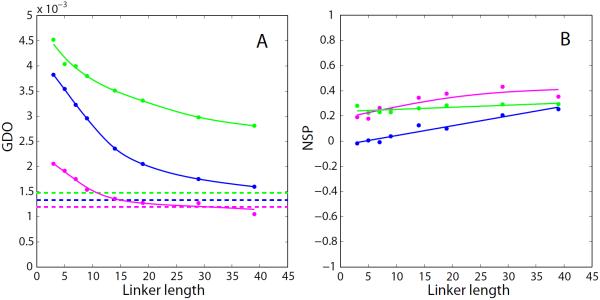
Fig. 3.
PALES simulations of steric alignment in a series of computer-generated tandem proteins. To generate structural ensembles, the two identical domains (represented by crystallographic or NMR coordinates and presumed to be fully rigid) were directly connected via glycine/serine linkers. The simulated couplings from all non-proline residues in the N-terminal domain were used to derive the alignment parameters. Other details of the structure generation and RDC simulations are as described in the Materials and Methods. Shown are the results for the tandem constructs of the disintegrin domain (structure 1MPZ,88 41 residue, radius of gyration 8.8 Å, magenta symbols), SH2 domain (extracted from structure 1A81,89 106 residues, 13.0 Å, blue), and nucleotide-binding domain (3GWI,90 170 residues, 15.3 Å, green). (a) Simulated GDO values for the domains which belong to the tandems (circles) and the corresponding isolated domains (dashed lines). (b) Simulated NSP values characterizing the mutual projection of the alignment tensors from (i) the domain which belongs to the tandem and (ii) the corresponding isolated domain. The spline curves are added to the graphs for visual guidance. To verify the convergence of the simulations, we have regenerated this plot using a different set of randomly generated structural ensembles. The results are shown in Fig. S1.