Tumours have high energetic and anabolic needs for rapid cell growth and proliferation1, and the serine biosynthetic pathway was recently identified as an important source of metabolic intermediates for these processes2,3. We integrated metabolic tracing and transcriptional profiling of a large panel of non-small cell lung cancer (NSCLC) cell lines to characterize the activity and regulation of the serine/glycine biosynthetic pathway in NSCLC. Here, we show that the activity of this pathway is highly heterogeneous and is regulated by NRF2, a transcription factor frequently deregulated in NSCLC. We found that NRF2 controls the expression of the key serine/glycine biosynthesis enzymes PHGDH, PSAT1 and SHMT2 via ATF4 to support glutathione and nucleotide production. Moreover, we show that expression of these genes confers poor prognosis in human NSCLC. Thus, a substantial fraction of human NSCLC activates a NRF2-dependent transcriptional program that regulates serine and glycine metabolism and is linked to clinical aggressiveness.
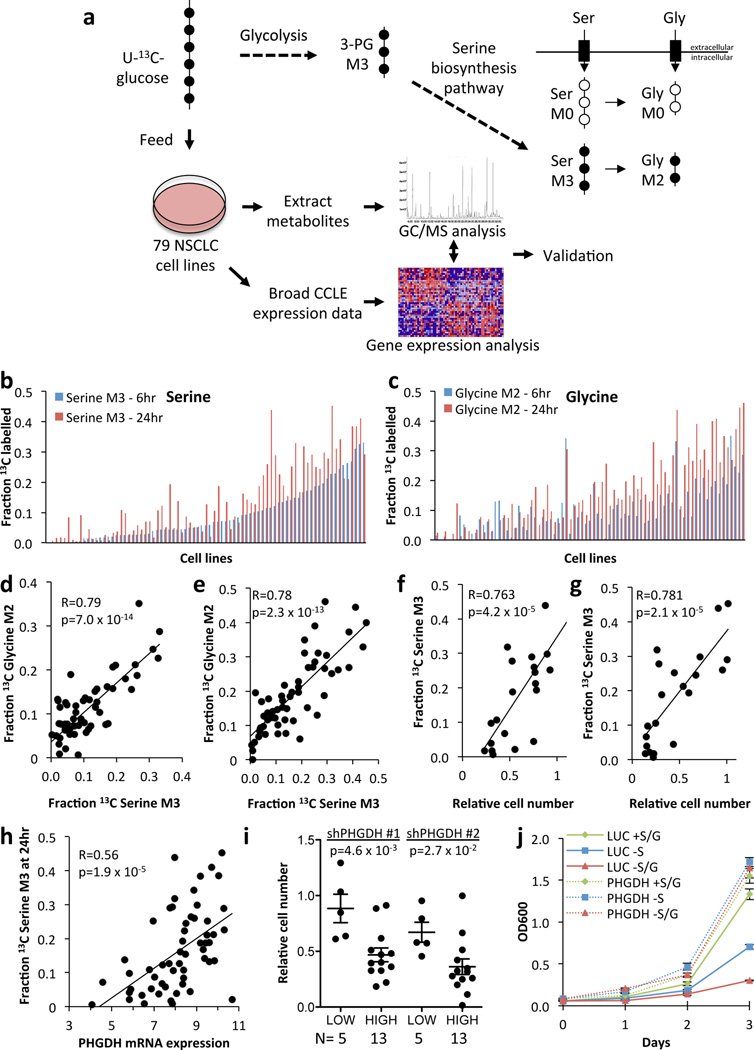
Uniformly labelled 13C-glucose ([U-13C] glucose) is metabolized via the glycolytic intermediate 3-phosphoglycerate to serine M3 (three 13C-labelled atoms) and, subsequently, to glycine M2 (two 13C-labelled atoms) through the combined action of the serine biosynthetic pathway and serine hydroxymethyltransferase4 (Fig. 1a). To profile the activity of the serine/glycine biosynthesis pathway in NSCLC, we labelled a panel of 79 human NSCLC cell lines with 13C-glucose and quantified serine and glycine labelling via gas chromatography/mass spectrometry (GC/MS). We determined 6 and 24 hours to be the optimal time points to detect labelling in serine and glycine (Supplementary Fig. 1). After 24 hours, the fractional abundance of serine M3 and glycine M2 ranged from 0–40% (Fig. 1b,c), with significant correlation between the time points (Supplementary Fig. 2). Neither serine nor glycine labelling correlated with cellular doubling times (Supplementary Fig. 3). There was a significant correlation between serine M3 and glycine M2 labelling at both time points, indicating that the glycine produced from glucose was derived from serine (Fig. 1d,e). As has been reported previously3, de novo serine synthesis conferred the ability to grow in the absence of extracellular serine (Fig. 1f,g). Thus, the serine biosynthesis pathway is not uniformly operant in NSCLC, and regulatory mechanisms exist for controlling the activation of this pathway in a subset of cell lines.
Figure 1.
Serine biosynthesis activity in lung cancer. (a) Synthesis of serine and glycine from glucose. Glycolytic metabolism of uniformly labelled 13C-glucose (U-13C-glucose) produces 3-phosphoglycerate (3-PG) labelled on all 3 carbons (M3). The serine biosynthetic pathway produces serine (M3, 3 carbons labelled), and subsequently glycine (M2, 2 carbons labelled) from 3-PG. Unlabelled (M0) serine and glycine are contributed from the cellular media. (b, c) Fraction of labelled intracellular serine (b) and glycine (c) at 6 and 24 hours in NSCLC cell lines. Cell lines are ordered according to serine M3 labelling at 6 hours. (d, e) Correlation between serine and glycine labelling at 6 (d) and 24 (e) hours. R=Pearson correlation coefficient. p-values were calculated by Student’s t-distribution with n–2 degrees of freedom. (f–g) Serine 'high' cells (serine M3 Z-score > 0.5 at 24 hours) are resistant to serine (f) or serine/glycine (g) starvation for 3 days. Each data point represents a cell line. (h) Serine labelling at 24 hours correlates with PHGDH mRNA expression. mRNA expression data was obtained from the CCLE. Each data point represents a cell line. (i) Serine ‘high’ cell lines demonstrate sensitivity to PHGDH knockdown that is not rescued by serine. Relative cell number was quantified 5 days after PHGDH knockdown. Each data point represents a cell line. (j) H1975 cells expressing luciferase (LUC) or PHGDH were assayed as in (f–g). Results are the average of 3 biological replicates. Error bars represent SEM here and for all figures.
To reveal a mechanism for increased serine biosynthesis in NSCLC, we correlated serine and glycine biosynthesis with gene expression using data from the Broad-Novartis Cancer Cell Line Encyclopedia5. We observed a significant correlation between serine and glycine labelling and the expression of PHGDH (Fig. 1h, Supplementary Table 1), the first and rate-limiting step in serine biosynthesis4. “Serine-high” cells (serine M3 Z-score > 0.5 at 24 hours) were sensitive to PHGDH silencing (Fig. 1i) and ectopic PHGDH rescued the proliferation of serine-low cell lines in serine-deficient media (Fig. 1j, Supplementary Fig. 4). Previous studies have revealed a role for PHGDH copy number gain in increasing serine biosynthetic activity2,3. However, we did not see evidence for this mechanism in NSCLC cell lines (Supplementary Fig. 5 and Supplementary Table 2). To investigate alternative mechanisms of PHGDH regulation, we performed gene set enrichment analysis (GSEA6,7) on the genes that positively correlated with serine and glycine biosynthesis. Interestingly, targets of the transcription factor nuclear factor erythroid-2–related factor 2 (NRF2, gene name NFE2L2) were the top hit (Supplementary Fig. 6), suggesting that NRF2 may be a regulator of PHGDH and the serine biosynthetic pathway.
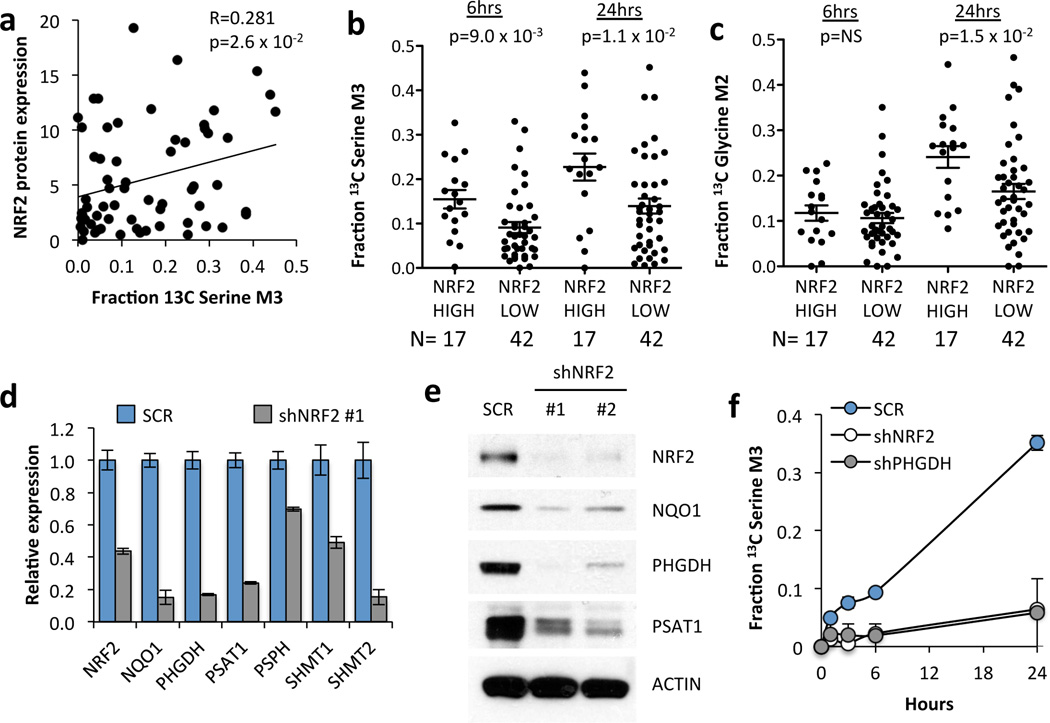
Next, we examined NRF2 localization (Supplementary Fig. 7) and found a significant correlation of nuclear NRF2 with serine biosynthesis (Fig. 2a). Additionally, we ranked these cell lines according to expression of NRF2 target genes (Supplementary Fig. 8 and Supplementary Table 3), which significantly correlated with NRF2 abundance (Supplementary Fig. 7). NRF2-high cell lines had significantly higher serine M3 labelling at 6 and 24 hours, and glycine M2 labelling at 24 hours (Fig. 2b,c). Next, we silenced NRF2 and found that it regulates serine/glycine biosynthetic pathway gene expression (PHGDH, PSAT1, PSPH, SHMT1, and SHMT2) (Fig. 2d and Supplementary Fig. 9). Decreased PHGDH and PSAT1 mRNA was accompanied by lower protein levels (Fig. 2e). Furthermore, NRF2 or PHGDH knockdown decreased the production of 13C-serine from 13C-glucose (Fig. 2f, Supplementary Fig. 10). These results demonstrate that transcriptional regulation of serine biosynthetic enzymes by NRF2 controls the production of serine from glucose.
Figure 2.
NRF2 regulates serine biosynthesis. (a) Nuclear NRF2 protein expression correlates with 13C-serine M3 labelling at 24 hours. Nuclear NRF2 protein expression data is found in Supplementary Fig. 7. (b) Cell lines with high NRF2 activity have significantly higher 13C serine M3 labelling at 6 and 24 hours. Cell lines were grouped according to the NRF2 score into “high” (>1.4) and “low” (<1.4). (c) NRF2 high cell lines have significantly higher 13C glycine M2 labelling at 24 hours. (b,c) p-values were calculated using an unpaired, two-tailed student’s t-test. (d) mRNA expression in A549 cells expressing scramble shRNA (SCR) or NRF2 shRNA #1. Decreased NQO1 expression confirmed that NRF2 activity was reduced upon knockdown. Results are the average of 3 technical replicates. (e) Western blot of serine pathway enzyme expression in lysates from A549 cells expressing scramble (SCR), or NRF2 shRNAs #1 or #2. (f) Cell lines were grown in the presence of U-13C-glucose for the indicated time points, the metabolites extracted and the fractional 13C-labelling on serine analysed by LC/MS. Results are the average of 3 biological replicates.
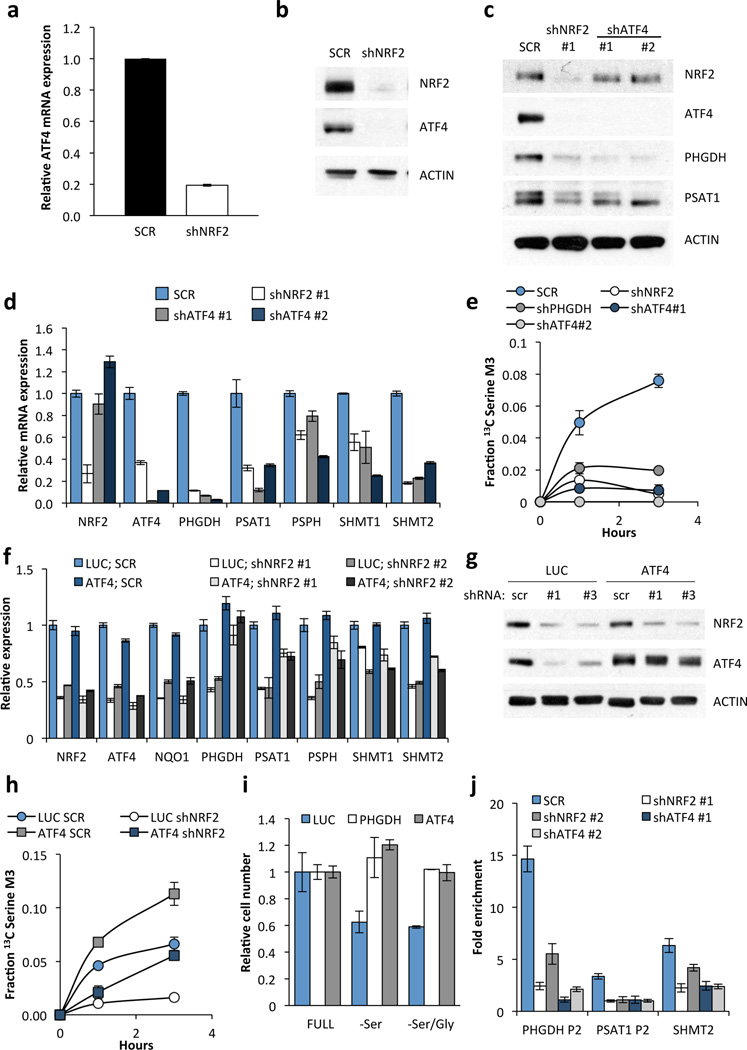
ATF4 transcriptionally activates serine biosynthetic genes in response to serine starvation in NSCLC cells8. Interestingly, nuclear ATF4 expression correlated with both serine labelling and NRF2 protein expression (Supplementary Fig. 11). ATF4 has been reported as both a direct transcriptional target9,10 and heterodimerization partner11,12 of NRF2. In agreement with transcriptional regulation, we observed a marked reduction in ATF4 mRNA expression, binding of RNA polymerase II to the ATF4 promoter, and levels of newly synthesized ATF4 mRNA upon NRF2 silencing (Fig. 3a, Supplementary Figs. 12a, 13a,b), with no effect on ATF4 mRNA stability (Supplementary Fig 13c,d). Furthermore, ATF4 protein was decreased upon NRF2 knockdown (Fig. 3b), but NRF2 did not regulate ATF4 translation13 (Supplementary Fig. 14). Notably, ATF4 knockdown reduced the expression and activity of the serine pathway to an extent similar to that seen with NRF2 knockdown, while it did not affect NRF2 expression (Fig. 3c–e). Ectopic expression of NRF2 in NRF2-depleted cells partially rescued NQO1, ATF4 and serine biosynthesis enzyme expression (Supplementary Fig. 12c) and induced expression of these genes in the serine-low cell line H1975 (Supplementary Fig. 12d), confirming the regulation of these genes by NRF2. Similarly, ectopic ATF4 expression rescued the effects of NRF2 silencing on serine biosynthesis enzyme expression (Fig. 3f,g, Supplementary Fig. 12b), and serine labelling from glucose at early time points (Fig. 3h), although defects in serine production were observed later (Supplementary Fig. 15). Furthermore, ectopic ATF4 rescued the growth of H1975 cells in serine deficient media (Fig. 3i). We identified the ATF4 binding sites in the PHGDH, PSAT1 and SHMT2 promoters (Supplementary Fig 16), and found that, while NRF2 itself did not bind to these sites (Supplementary Fig. 16), its silencing significantly decreased binding of ATF4 (Fig. 3j). These results demonstrate that NRF2 regulates serine biosynthesis gene expression through ATF4.
Figure 3.
NRF2 regulates the expression of serine/glycine biosynthesis genes through ATF4. (a) ATF4 mRNA expression in A549 cells expressing scramble shRNA (SCR), or NRF2 shRNA #1. (b) Western blot of NRF2, ATF4 and ACTIN expression in cells from (a). (c) Western blot of NRF2, ATF4 and serine pathway enzyme expression in lysates from A549s expressing scramble (SCR), NRF2 shRNA #1, or ATF4 shRNAs #1 or #2. (d) mRNA expression in cells from (c). (e) ATF4 knockdown impairs serine biosynthesis. Cell lines from were grown in the presence of U-13C-glucose for the indicated time points, the metabolites extracted and the fractional 13C-labeling on serine analysed by LC/MS. (f) ATF4 rescues serine biosynthesis enzyme expression following NRF2 knockdown. A549 cells were infected with lentivirus encoding mATF4 prior to infection with scramble or NRF2-targeting lentivirus. (g) Western analysis of NRF2, ATF4, and ACTIN expression in the cells from (f). (h) ATF4 rescues the serine biosynthesis defect in shNRF2 A549 cells. Cells were assayed as in (e). (i) ATF4 rescues the growth of H1975 cells in serine deficient media. Cells expressing luciferase (LUC) or ATF4 were grown in the indicated media for 3 days and cell number normalized to cells grown in full media. (j) Chromatin immunoprecipitation of ATF4 to the PHGDH, PSAT1 and SHMT2 promoters. Samples were normalized to IgG control immunoprecipitations. Results are the average of 3 technical (a, d, f, j) or biological (e, h, i) replicates.
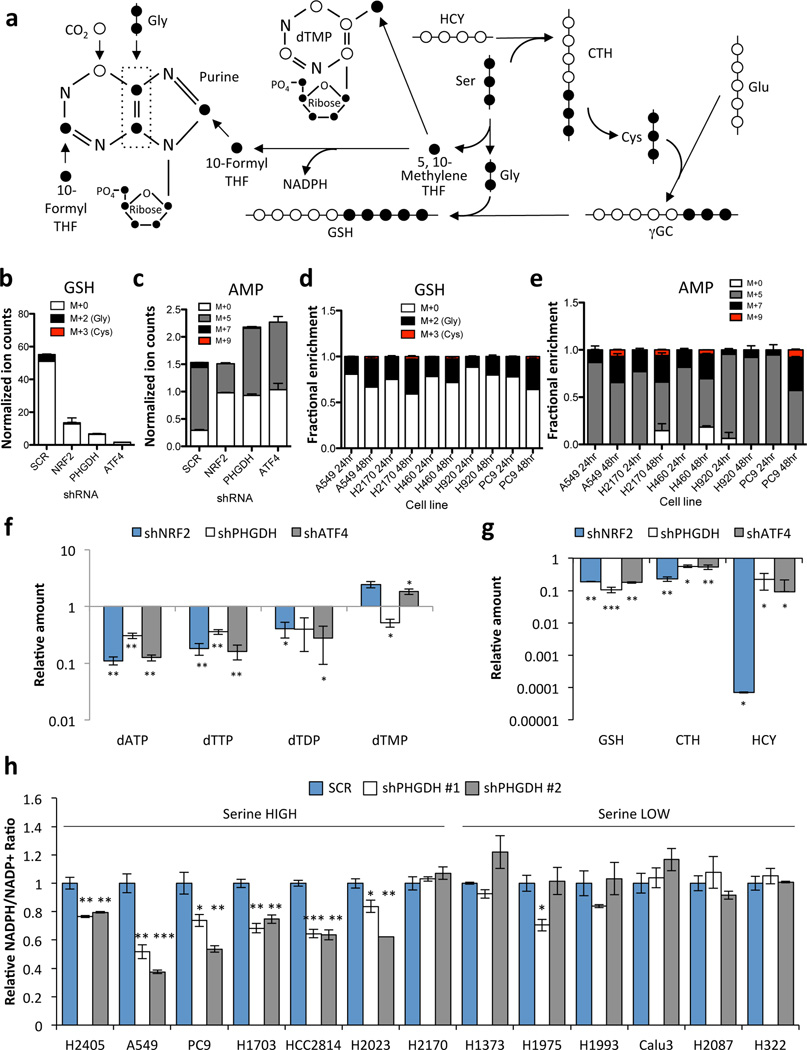
We examined how PHGDH-derived serine contributes to downstream metabolism (Fig. 4a). PHGDH, NRF2 or ATF4 silencing decreased the incorporation of glucose-derived serine into cystathionine (Supplementary Fig. 17a,b), and glucose-derived glycine into glutathione (Fig. 4b, Supplementary Fig. 17c), without loss of cell viability (Supplementary Fig. 18). Metabolism of serine to glycine results in the production of one-carbon units via the folate cycle that are utilized for purine and thymidine synthesis (Fig. 4a). We observed a decrease in the PHGDH-derived labelling (M+7) of purines including IMP, AMP, ADP, and inosine following PHGDH or ATF4 silencing, while NRF2 silencing decreased both the ribose (M+5) and PHGDH-derived (M+7) labelling of these purines (Fig. 4c, Supplementary Fig. 17d–f). Serine-high cell line labelling in glutathione and purines was significantly higher at 48 hours than 24 hours (Fig. 4d,e, Supplementary Fig. 19a,b). Furthermore, the majority of the 13C-labelled serine and glycine had escaped the cell by 24 hours of 13C-glucose labelling due to equilibration with unlabelled amino acids in the media, suggesting that the fraction labelling at 24 hours was underestimating the total contribution of PHGDH to these metabolite pools (Supplementary Fig. 20, Supplementary Discussion). In support of this notion, PHGDH, NRF2 or ATF4 silencing resulted in significant decreases in the total levels of purines and thymidine nucleotides (Fig. 4f) as well as glutathione, cystathionine and homocysteine (Fig. 4g). In contrast, we did not observe differences in the S-adenosyl methionine (SAM)/S-adenosyl homocysteine (SAH) ratio, which is also modulated by the folate cycle, or in histone or DNA methylation (Supplementary Fig. 21). Furthermore, we observed a significant decrease in the NADPH/NADP+ ratio in serine-high cells following PHGDH silencing that was not observed in serine-low cells (Fig. 4h). These results demonstrate that the serine biosynthesis pathway supports glutathione and nucleotide production in NSCLC.
Figure 4.
PHGDH-derived serine supports the transsulfuration and folate cycles. (a) Serine metabolism via the transsulfuration and folate cycles. 13C-labelled carbons (●) unlabelled carbons (○). Gly: Glycine, Ser: Serine, CTH: Cystathionine; HCY: Homocysteine, GSH: Glutathione, γGC: γ-glutamyl cysteine, Glu: Glutamate. (b–e) A549 cells expressing scramble (SCR), NRF2 or PHGDH shRNAs were grown in the presence of U-13C-glucose for 24 hours and metabolites were extracted and analysed by LC/MS. (b) Analysis of 13C-labelling on the glycine component of glutathione. (c) Analysis of 13C-labeling on the purine metabolite adenosine monophosphate (AMP). (d,e) Analysis of glutathione (d) and AMP (e) labelling in serine high cell lines. Cells were labelled with 13C-glucose for 24 or 48 hours as indicated. (b,d) PHGDH-derived serine is incorporated into glutathione through the generation of glycine and cysteine. M+0 denotes no carbons labelled, M+2 denotes labelling on the glycine moiety, M+3 denotes labeling on the cysteine moiety, and M+5 denotes labeling on both glycine and cysteine. M+5 labeling was not observed. (c,e) M+5 labelling occurs following ribose-5-phosphate labelling via the pentose phosphate pathway. M+7 labelling is the result of ribose labelling plus either glycine or formyl-THF labelling in the purine ring. M+9 labelling is the result of ribose labelling plus either glycine and formyl-THF labelling in the purine ring. M+0 has no labelled carbons. (f–g) LC/MS analysis of total metabolite levels in the nucleotide (f) and transsulfuration (g) pathways. (h) NADPH/NADP+ ratios 4 days after PHGDH knockdown. Results are the average of 3 biological replicates.
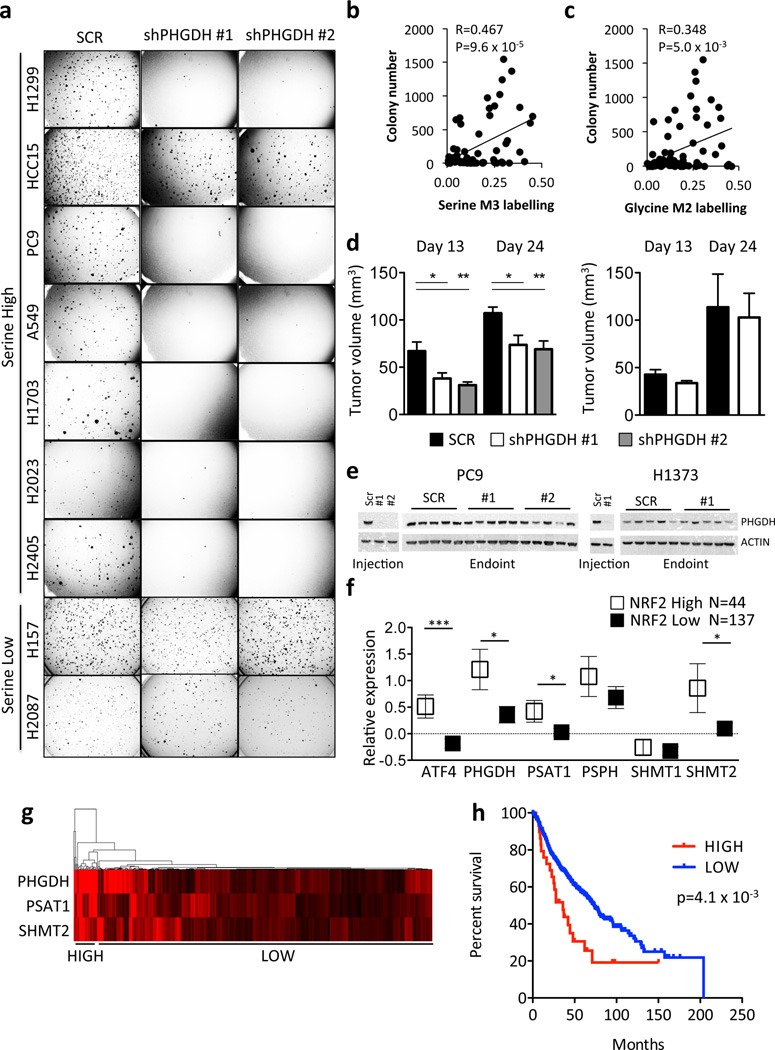
We next asked whether this pathway promotes tumourigenesis. PHGDH silencing significantly impaired the soft agar growth of serine-high, but not serine-low, cell lines (Fig. 5a). Interestingly, we observed a significant correlation between serine and glycine labelling and colony number in soft agar (Fig. 5b,c). Furthermore, PHGDH silencing impaired the xenograft growth of the serine-high cell line PC9 (Fig. 5d, left) but not the serine-low cell line H1373 (Fig. 5d, right). We observed that all tumours re-expressed PHGDH at endpoint (Fig. 5e), suggesting that PHGDH was required for tumour formation. Next, we examined whether serine pathway gene expression correlated with overall survival of human patients. Human tumours with high NRF2 protein expression displayed significantly higher expression of ATF4, PHGDH, PSAT1, and SHMT2 mRNA (Fig. 5f), and vice versa (Supplementary Fig. 22a). Furthermore, we found that high expression of PHGDH, PSAT1 and SHMT2 conferred a significantly poorer prognosis (Fig. 5g,h) and was associated with higher tumour grade (Supplementary Fig. 22b–d). These results demonstrate that in human NSCLC, NRF2 regulates the expression of serine biosynthetic enzymes, which correlates with poor prognosis.
Figure 5.
Activation of the serine biosynthesis pathway promotes tumourigenesis in NSCLC. (a) PHGDH knockdown impairs soft agar growth of serine high, but not serine low, cell lines. (b–c) Soft agar growth correlates with serine (b) and glycine (c) labelling at 24 hours. Each cell line was plated at 5,000 cells/well and the number of colonies counted after 14 days. (d) PHGDH knockdown impairs the xenograft growth of a serine high cell line (PC9, left) but not a serine low cell line (H1373, right). Results are the average of 5 tumors. (e) Western analysis of PHGDH expression of cell lines and xenografts from (d) upon injection and at endpoint. (f) Patients with high NRF2 protein expression (Z-score > 0.5) demonstrate elevated serine pathway gene expression in patient samples from TCGA lung adenocarcinoma cohort. Boxes represent mean values, error bars represent SEM. (g) Gene expression of PHGDH, PSAT1 and SHMT2 in the Director’s Consortium lung adenocarcinoma dataset clusters patients into high and low expression cohorts. (h) Kaplan-Meier survival analysis of patients with high (n = 29, red) or low (n = 414, blue) expression of PHGDH, PSAT1 and SHMT2 based on the patient clustering from (g). Median survival is 36 (high) vs. 73.2 (low) months. The p-value was calculated using the Mantel-Cox test.
We have demonstrated a striking heterogeneity in the activity of the serine biosynthetic pathway in NSCLC. Notably, intracellular amino acid labelling from 13C-glucose is underestimated because of rapid exchange with 12C-amino acids from the media, which is likely mediated by amino acid antiporters (see Supplementary Note). Caution should be used when interpreting data from 13C-labelling experiments due to these exchange mechanisms.
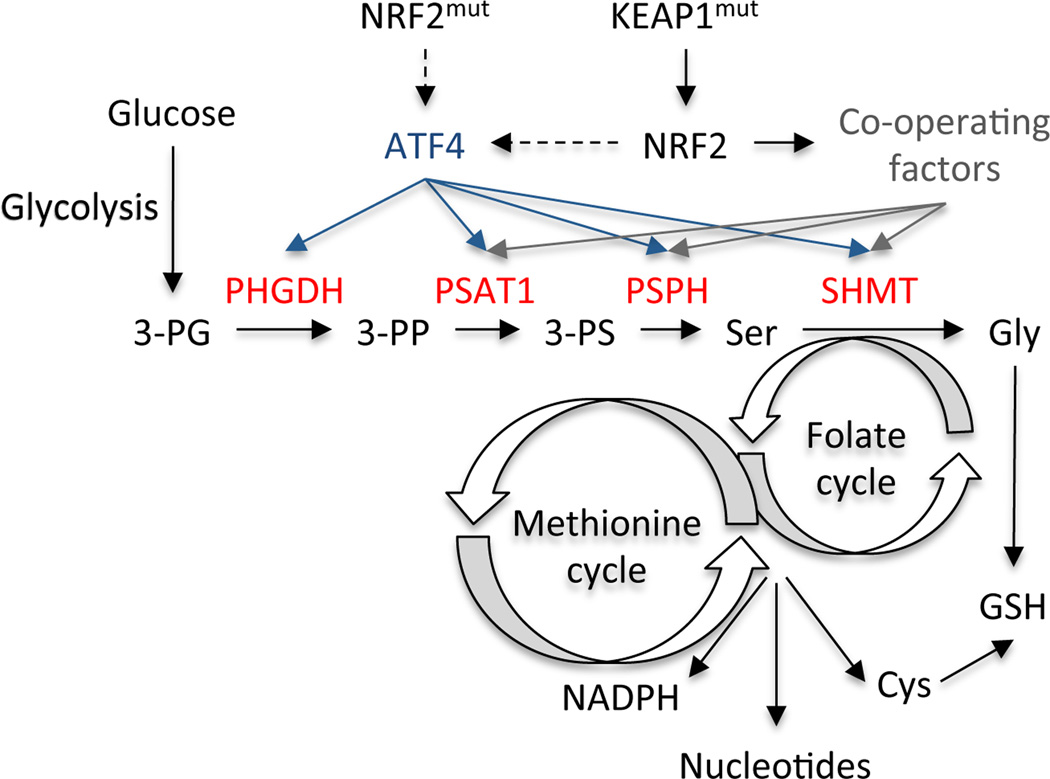
Heterogeneity of metabolic or signalling pathways is a common phenomenon across tumours, cell lines, and even between cells of the same tumour. Here, by systematically analysing the serine/glycine biosynthesis pathway in a large, highly-annotated panel of NSCLC cell lines, we identified NRF2 as the molecular driver of this pathway. NRF2 is frequently deregulated in NSCLC through somatic mutations that disrupt the NRF2–KEAP1 interaction to constitutively activate NRF214–18. Cancers with high NRF2 levels are associated with poor prognosis19,20, resistance to therapy and rapid proliferation18,21. Importantly, NRF2 ablation in various tumour models results in elevated ROS and the suppression of tumour growth in vivo21–23. However, the precise mechanism by which NRF2 promotes tumourigenesis is unclear. Recent studies have revealed that, in addition to genes that promote ROS detoxification, NRF2 regulates genes involved in anabolic metabolism24,25. Here, we demonstrate that NRF2 regulates a serine biosynthesis metabolic program via ATF4 and PHGDH to supply the substrates for glutathione and nucleotide production (Figure 6), which synergizes with the regulation of the pentose phosphate pathway to supply ribose for nucleotides24,25. NRF2 indirectly regulates ATF4 transcription via unknown mechanisms. Additionally, while ATF4 completely rescues PHGDH expression following NRF2 depletion, the partial rescue of other serine biosynthesis genes suggests that NRF2 regulates these genes combinatorially through ATF4 and additional factors. Furthermore, these findings suggest that multiple NRF2-regulated pathways co-ordinately contribute to tumourigenesis. Our work encourages the integration of metabolite tracing on large panels of cancer cell lines in combination with gene expression analysis. This approach is a powerful tool for determining mechanisms responsible for differential regulation of metabolic pathways and may reveal additional links between the activity of metabolic pathways and genetic alterations in cancers.
Figure 6.
Model of the regulation of serine/glycine biosynthesis by NRF2. An ATF4 transcriptional program, indirectly activated by NRF2/KEAP1 mutations, regulates the expression of serine/glycine biosynthesis enzymes. These enzymes produce serine and glycine from the glycolytic intermediate 3-PG and funnel the carbon into glutathione and nucleotides via the folate and transsulfuration cycles. NRF2/ATF4 regulated enzymes are shown in red. 3-PG: 3-phosphoglycerate; 3-PP: 3-phosphohydroxy pyruvate; 3-PS: 3-phosphoserine; Ser: Serine; Cys: Cysteine; GSH: Glutathione.
Online Methods
Animals
Male nude mice (CrTac:NCr-Foxn1nu) were obtained from Taconic labs and maintained under pathogen-free conditions. Experiments were performed according to IACUC guidelines. Mice were injected at 8 weeks of age with 2×106 NSCLC cells on each flank. shRNAs were randomized so they were evenly distributed across mice. As the animal study was exploratory, no statistical test was used to determine adequate sample size. No mice were excluded from the analysis. The study was not blinded.
Cell culture
All NSCLC cell lines used in this study were obtained from the Hamon Cancer Center Collection (University of Texas Southwestern Medical Center). Cells were maintained in RPMI-1640 (Life Technologies Inc.) supplemented with 5% or 10% fetal calf serum (FCS) without antibiotics at 37°C in a humidified atmosphere containing 5% CO2 and 95% air. All experiments were performed in media containing serine and glycine except where otherwise noted. All cell lines have been DNA fingerprinted using the PowerPlex 1.2 kit (Promega) and mycoplasma tested by the e-Myco (Boca Scientific) or MycoAlert kit (Lonza). Although NCI-H157 is listed in the database of commonly misidentified cell lines, it was originally derived by J.D.M. and fingerprinted before use.
Celltox green staining
Cells were incubated in RPMI + 10% FCS containing 1× Celltox green (Promega) and 5 uM Syto 17 (Life Technologies) for 20 minutes, washed in PBS, and imaged on an EVOS FL cell imaging system (Life Technologies).
Chromatin Immunoprecipitation
Cells (5×106) were fixed at 37°C in RPMI with 1% formaldehyde for 10 min, lysed in 1% SDS, 10 mM EDTA, 50 mM Tris-HCl pH 8.1 plus protease inhibitors and sonicated with a probe tip until DNA was an average size of 1 kilobase. Input was saved and lysate was diluted in immunoprecipitation buffer (1% Triton, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl pH 8.1) and mixed with beads (Dynal Protein A, Invitrogen) that were pre-bound overnight with antibodies to Nrf2 (H-300, Santa Cruz), ATF4 (11815, Cell Signaling), RNA pol II (pSer5, ab5131, abcam) or Rabbit IgG (sc-2027, Santa Cruz). Chromatin was immunoprecipitated overnight, and beads were washed six times with RIPA buffer (50 mM HEPES pH 7.6, 1 mM EDTA, 0.7% Na deoxycholate, 1% NP-40, 0.5 M LiCl) and twice with TE. Beads were incubated with 1% SDS, 0.1 M NaHCO3 for 30 min at room temperature, and then crosslinks were reversed on both the input and the immunoprecipitate by heating overnight in a 65°C water bath. DNA was purified with a QIAquick spin kit (Qiagen) and Q-PCR was performed in triplicate with a Fast Sybr green master mix on a Step One Real-Time PCR system (all Life Technologies). Primer sequences can be found in Supplementary Table 4.
DNA methylation
DNA was extracted from cells in lysis buffer (10 mM Tris, 100 mM NaCl, 10 mM EDTA, 0.5% SDS, 0.4 µg/ml Proteinase K, pH 8.0) by heating overnight at 65°C. DNA was purified by precipitating protein with NaCl, and precipitated with isopropanol. 2 ug DNA was denatured in 0.4 M NaOH, 10 mM EDTA at 95°C for 10min and neutralized with an equal volume of 2 M ammonium acetate, pH7.0. 200 ng of DNA was spotted on a nylon membrane (GE Healthcare), cross linked at maximum twice, and blocked in 5% milk for 1 hour. The membrane was incubated overnight with 5-meC antibody (clone 33D3; 1:1,000; Epigentek), followed by an HRP-secondary and chemiluminescence visualization. To ensure equal loading, the membrane was stained with 0.02% methylene blue in 0.3 M sodium acetate, pH 5.2. 5-meC intensity was normalized to methylene blue.
GC/MS metabolite tracing
All NSCLC cell lines were cultured under identical conditions to identify cell-autonomous differences in glucose metabolism. Cells were seeded into 60-mm culture dishes and grown until 70–80% confluent. Each dish was then rinsed in warm phosphate-buffered saline and overlaid with 4 mL of RPMI-1640 containing 5% dialyzed fetal calf serum, 4 mM unlabelled glutamine, and 10 mM [U-13C] glucose (Cambridge Isotope Laboratories). NB: RPMI-1640 contains unlabelled serine and glycine. After 6 hours or 24 hours, the medium was aspirated and cells were rapidly rinsed in cold normal saline solution. The cells were lysed in 0.5 mL of cold 50% methanol, with three rapid cycles of freeze-thawing between liquid nitrogen and a 37°C water bath. The lysates were cleared of cellular debris by centrifugation, and metabolites in the supernatant were evaporated under blown air or by centrifugation under vacuum. Derivatization, mass spectrometry, and mathematical correction for natural abundance isotopes were performed according to published methods26,27. The following fragments were monitored, which represent derivatized species: serine – m/z 306 (M+0) – 309 (M+3); glycine, m/z 276 (M+0) – 278 (M+2). Every cell line was analysed in biological replicates where n≥3 and no two replicates were performed on the same day.
Lentivirus production and infection
Lenti-X 293T cells (Clontech) were transfected at 90% confluence with Lipofectamine 2000 (Invitrogen). Packaging plasmids pCMV-dR8.2 dvpr (addgene # 8455) and pCMV-VSV-G (addgene #8454) were used with the vectors found in Supplementary Table 5.
Viral supernatants were collected at 48 and 72 hours and added to target cells with 8µg/ml polybrene for 3 hours. Cells were selected in 1 µg/ml puromycin where appropriate.
LC/MS measurement of total and 13C-labeled metabolites in A549 cells. Cells were plated the day before labelling at 2×106 cells/10 cm dish and media was changed to glucose-free RPMI containing 10% dialyzed serum + 10 mM U-13C-glucose (Cambridge Isotope Laboratories) for the indicated time points. Metabolites were extracted in ice-cold 80% methanol and analysed by targeted LC-MS/MS via selected reaction monitoring (SRM), as described28. The following precursor ions were monitored: serine – m/z 106 (M+0) and 109 (M+3); cystathionine – m/z 223 (M+0) and 226 (M+3); glutathione – m/z 308 (M+0), glutathione (G) – m/z 310 (M+2); homocysteine – m/z; 136; IMP – m/z 349 (M+0), 354 (M+5) and 356 (M+7); inosine – m/z 267 (M+0), 272 (M+5), 274 (M+7); AMP – m/z 348 (M+0), 353 (M+5) and 355 (M+7); dATP – m/z 490; dTTP – m/z 481; dTDP – m/z 401; dTMP – m/z 323. The bracketed amino acid following glutathione indicates which constituent amino acid is 13C labelled. Samples were analysed in triplicate. Data represents median-normalized values.
Luciferase assays
Luciferase assays were performed with the Dual-Glo Luciferase Assay System (Promega) according to the manufacturer’s instructions.
NADPH/NADP+ ratios
NADP+ and NADPH were determined with the NADP/NADPH-glo assay kit (Promega) according to the manufacturer’s protocol.
NRF2 score calculation
Cell lines were grouped into high and low groups based on the expression of five classic NRF2 target genes: NQO1, GCLC, GLCM, SLC7A11, and AKR1C1. The expression of each gene was normalized to the median across the cell lines, and these five genes were then averaged together. NRF2 high vs. low was defined as the top quartile vs. bottom three quartiles. The top 20 overexpressed genes in the NRF2 high cell lines compared to the NRF2 low were used to calculate the NRF2 score: AKR1C1, AKR1C2, SPP1, ALDH3A1, LOC344887, AKR1C3, OSGIN1, PGD, CYP4F11, AKR1B10, KIAA0319, SRXN1, NR0B1, SLC7A11, LOC100292680, ABCC2, CABYR, JAKMIP3, KYNU and PTGR1. Gene expression values were normalized to the median and then these 20 genes were averaged together to get the NRF2 score. Individual gene expression values and NRF2 scores can be found in Supplementary Table 3, and NRF2 high vs. low clustering is found in Supplementary Fig. 8.
Patient samples and survival analysis
The National Cancer Institute Director’s Challenge Consortium study (Director’s Consortium)29 and the Cancer Genome Atlas (TCGA) lung adenocarcinoma data were used in this study to evaluate gene signatures’ prognostic performance. The Director’s Consortium data set collected 442 resected lung adenocarcinomas at four US institutions29, while the TCGA Research Network data includes 203 patient samples for which gene expression and survival data are available. Unsupervised cluster analysis was used to group patients based on expression of PHGDH, PSAT1 and SHMT2, using average linkage clustering with the Spearman's rank correlation distance metric. Clustering was performed with Cluster 3.0. Heat map visualization was performed with JavaTreeView. Kaplan–Meier survival curves were used to determine survival rate as a function of time and survival differences were analysed by a log-rank Mantel-Cox test using Graph Pad Prism.
Proliferation assays
Cells were seeded at 500–10,000 cells/well in 96 well plates on day −1 and infected on day 0 with lentivirus. Alternatively, cells were switched into RPMI + 10% dialyzed FBS containing full amino acids, lacking serine, or lacking serine and glycine. Plates were fixed on the indicated days with 4% paraformaldehyde, stained with crystal violet, washed and dried. Crystal violet was solubilized in 10% acetic acid and the OD600 was measured.
Pulse-labelling with 4-thiouridine
Cells were labelled on 10 cm dishes at 70% confluence with 200 uM 4-thiouridine (4sU, Sigma Aldrich) for 60 minutes as described30. RNA was extracted with Trizol. 4-thiouridine containing mRNA molecules were biotinylated with biotin-HPDP (EZ-Link Biotin-HPDP, Pierce, Cat #21341), and purified with the uMACS streptavidin kit (Miltenyi Biotec). RNA was eluted in 100 mM DTT, cDNA was synthesized, and ATF4 levels quantified and normalized to β-actin.
Reagents
Serine and glycine were purchased from Sigma Aldrich. RPMI media lacking serine and glycine was custom prepared by Life Technologies.
Real-time PCR
RNA was isolated using an RNeasy kit (Qiagen). cDNA was synthesized using the Superscript VILO Master mix and analysed by quantitative PCR (q-PCR) using Fast Sybr green master mix on a Step One Real-Time PCR system (all Life Technologies). Target gene expression was normalized to actin expression, and shown relative to control samples. Primer sequences can be found in Supplementary Table 4.
siRNA transfection
100,000 cells/well were reverse transfected in 800 ul of growth media in 12 well dishes. 2 ul DharmaFECT Duo was combined with 100 pmol siRNA in a final volume of 200 ul according to the manufacturer’s instructions, which was then added to the cells. Cells were analysed after 2 days. Dharmacon ON-TARGET plus non-targeting siRNA (D-001810-10) and NFE2L2 (L-003755-00-0005) pools were used.
Soft agar assays
Soft agar assays were performed in triplicate in 6-well dishes. A 1 mL base layer of 0.8% agar in RPMI was plated and allowed to solidify, then 5,000 cells/well were plated in 0.4% agar on top. 1mL of RPMI was added the following day to each well, and changed as needed. NB: RPMI contains serine and glycine. Soft agar was stained with 0.01% crystal violet in 4%PFA/PBS solution and imaged in a ChemiDoc system (Bio-Rad). Colonies were quantified with Image J.
Statistical analysis
Data were analysed using a two-sided unpaired Student’s t test and the Mantel-Cox test was used for survival analyses. For all statistical analyses GraphPad Prism 6 software was used, and values of p < 0.05 were considered statistically significant (*P < 0.05; **P < 0.01; ***P < 0.001). The mean ± standard error of the mean of at least three independent experiments performed in triplicates is reported. For all experiments similar variances between groups were observed. Normal distribution of samples was not determined.
Western blotting
Protein lysates were prepared using RIPA lysis buffer and separated on 4–12% NuPAGE gels (Invitrogen), transferred onto a nitrocellulose membrane (Millipore) and incubated with the following antibodies: ACTIN (ab6276) and NRF2 (EP1808Y) mAb (both Abcam), NQO1 (HPA007308) and PHGDH (HPA021241) (both Sigma), PSAT1 (PA5-22124, Pierce), and ATF4 (11815, Cell Signaling). Alternatively, nuclear extracts were prepared as described31. Histone extracts were prepared with the Histone Extraction Kit according to the manufacturer’s instructions (Abcam, ab113476). Histone extracts were probed with the following antibodies: H3K4me3 (9727, Cell Signaling), H3K27me3 (07-449, Millipore), total H3 (4499, Cell Signaling).
Supplementary Material
Acknowledgements
We would like to thank George Poulogiannis for bioinformatics advice and Hansaa Abbasi, Claire Klimko and Min Yuan for technical support with mass spectrometry experiments. This work was supported by NIH grants P01 CA117969 and R01 GM041890 (L.C.C.), R01 CA157996-01 (R.J.D.), 5R01CA152301 (Y.X.), P50CA70907 (J.D.M, I.I.W., Y.X., K.E.H.), and CPRIT funding to J.D.M, Y.X., I.I.W., K.E.H. (RP110708, RP120732) and R.J.D (RP130272). P-H.C. was supported by a grant from the Welch Foundation to R.J.D. (I-1733). The mass spectrometry work was partially supported by NIH grants 5P30CA006516 and 5 P01CA120964 (J.M.A.). G.M.D. was the Malcolm AS Moore Hope Funds for Cancer Research Fellow and is supported by the PanCAN/AACR Pathway to Leadership grant.
Competing financial interests
L.C.C. owns equity in, receives compensation from, and serves on the Board of Directors and Scientific Advisory Board of Agios Pharmaceuticals. Agios Pharmaceuticals is identifying metabolic pathways of cancer cells and developing drugs to inhibit such enzymes in order to disrupt tumour cell growth and survival. R.J.D. is on the scientific advisory boards of Agios Pharmaceuticals and Peloton Therapeutics. Peloton Therapeutics is developing drugs to target altered molecular pathways in cancer, including altered metabolism.
Footnotes
URLs
the TCGA Research Network: http://cancergenome.nih.gov/
Author Contributions
G.M.D., R.J.D., and L.C.C. designed the study. G.M.D. ad E.M. performed molecular biology experiments. G.M.D., P.H.C., E.M., J.A.S., Z.H., and J.M.A. performed metabolomics and isotope labelling and analysed the data. D.W. performed xenograft experiments. H.T. and Y.X. performed bioinformatics analysis. K.E.H., I.I.W. and J.D.M. contributed highly annotated lung cancer cell lines. G.M.D., E.M., and L.C.C. wrote the manuscript. All authors commented on the manuscript.
References
- 1.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Locasale JW, et al. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet. 2011;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Possemato R, et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476:346–350. doi: 10.1038/nature10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mullarky E, Mattaini KR, Vander Heiden MG, Cantley LC, Locasale JW. PHGDH amplification and altered glucose metabolism in human melanoma. Pigment Cell Melanoma Res. 2011;24:1112–1115. doi: 10.1111/j.1755-148X.2011.00919.x. [DOI] [PubMed] [Google Scholar]
- 5.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 7.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye J, et al. Pyruvate kinase M2 promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation. Proc Natl Acad Sci U S A. 2012;109:6904–6909. doi: 10.1073/pnas.1204176109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyamoto N, et al. Transcriptional regulation of activating transcription factor 4 under oxidative stress in retinal pigment epithelial ARPE-19/HPV-16 cells. Invest. Ophthalmol. Vis. Sci. 2011;52:1226–1234. doi: 10.1167/iovs.10-5775. [DOI] [PubMed] [Google Scholar]
- 10.Afonyushkin T, et al. Oxidized phospholipids regulate expression of ATF4 and VEGF in endothelial cells via NRF2-dependent mechanism: novel point of convergence between electrophilic and unfolded protein stress pathways. Arterioscler Thromb Vasc Biol. 2010;30:1007–1013. doi: 10.1161/ATVBAHA.110.204354. [DOI] [PubMed] [Google Scholar]
- 11.Ye P, et al. Nrf2- and ATF4-dependent upregulation of xCT modulates the sensitivity of T24 bladder carcinoma cells to proteasome inhibition. Mol Cell Biol. 2014;34:3421–3434. doi: 10.1128/MCB.00221-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He CH, et al. Identification of activating transcription factor 4 (ATF4) as an Nrf2-interacting protein. Implication for heme oxygenase-1 gene regulation. J Biol Chem. 2001;276:20858–20865. doi: 10.1074/jbc.M101198200. [DOI] [PubMed] [Google Scholar]
- 13.Harding HP, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 14.Hayes JD, McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem Sci. 2009;34:176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Kim YR, et al. Oncogenic NRF2 mutations in squamous cell carcinomas of oesophagus and skin. J Pathol. 2010;220:446–451. doi: 10.1002/path.2653. [DOI] [PubMed] [Google Scholar]
- 16.Konstantinopoulos PA, et al. Keap1 mutations and Nrf2 pathway activation in epithelial ovarian cancer. Cancer Res. 2011;71:5081–5089. doi: 10.1158/0008-5472.CAN-10-4668. [DOI] [PubMed] [Google Scholar]
- 17.Seng S, et al. NRP/B mutations impair Nrf2-dependent NQO1 induction in human primary brain tumors. Oncogene. 2009;28:378–389. doi: 10.1038/onc.2008.396. [DOI] [PubMed] [Google Scholar]
- 18.Zhang P, et al. Loss of Kelch-like ECH-associated protein 1 function in prostate cancer cells causes chemoresistance and radioresistance and promotes tumor growth. Mol Cancer Ther. 2010;9:336–346. doi: 10.1158/1535-7163.MCT-09-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shibata T, et al. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci U S A. 2008;105:13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solis LM, et al. Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin Cancer Res. 2010;16:3743–3753. doi: 10.1158/1078-0432.CCR-09-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh A, et al. RNAi-mediated silencing of nuclear factor erythroid-2-related factor 2 gene expression in non-small cell lung cancer inhibits tumor growth and increases efficacy of chemotherapy. Cancer Res. 2008;68:7975–7984. doi: 10.1158/0008-5472.CAN-08-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeNicola GM, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohta T, et al. Loss of Keap1 function activates Nrf2 and provides advantages for lung cancer cell growth. Cancer Res. 2008;68:1303–1309. doi: 10.1158/0008-5472.CAN-07-5003. [DOI] [PubMed] [Google Scholar]
- 24.Mitsuishi Y, et al. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 25.Singh A, et al. Transcription factor NRF2 regulates miR-1 and miR-206 to drive tumorigenesis. J Clin Invest. 2013;123:2921–2934. doi: 10.1172/JCI66353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 26.Cheng T, et al. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc Natl Acad Sci U S A. 2011;108:8674–8679. doi: 10.1073/pnas.1016627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullen AR, et al. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan M, Breitkopf SB, Yang X, Asara JMA. positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc. 2012;7:872–881. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shedden K, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rädle B, et al. Metabolic labeling of newly transcribed RNA for high resolution gene expression profiling of RNA synthesis, processing and decay in cell culture. J Vis Exp. 2013 doi: 10.3791/50195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meylan E, et al. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature. 2009;462:104–107. doi: 10.1038/nature08462. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.