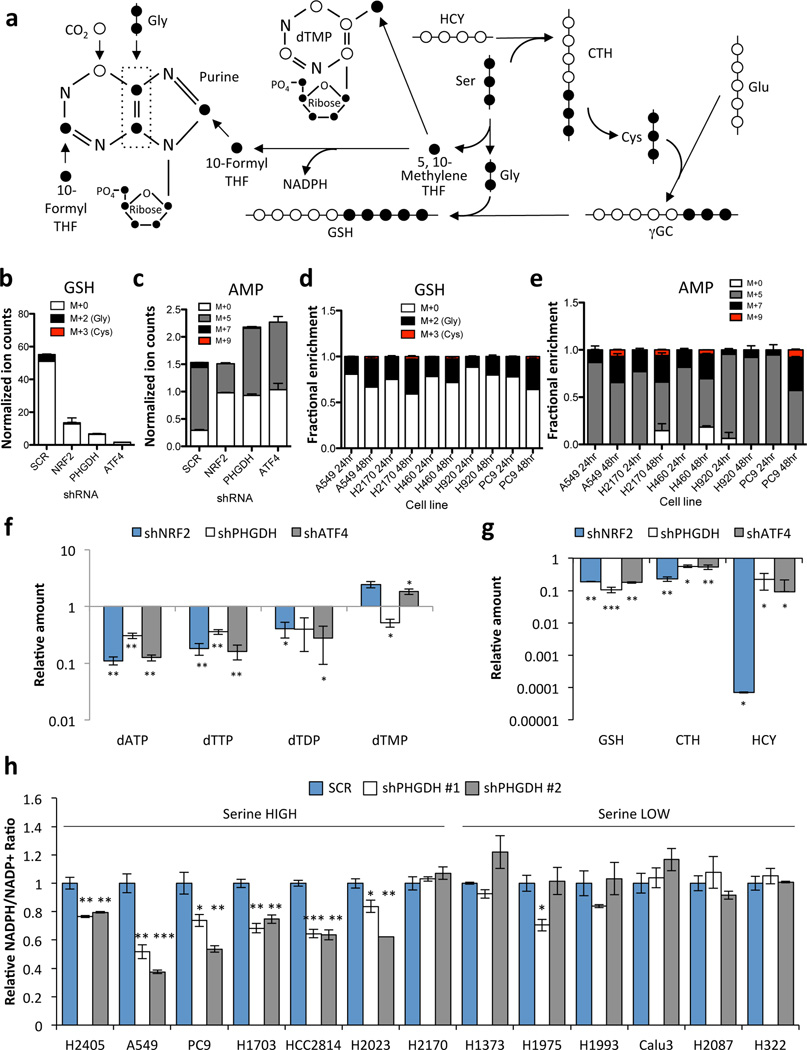
Figure 4.
PHGDH-derived serine supports the transsulfuration and folate cycles. (a) Serine metabolism via the transsulfuration and folate cycles. 13C-labelled carbons (●) unlabelled carbons (○). Gly: Glycine, Ser: Serine, CTH: Cystathionine; HCY: Homocysteine, GSH: Glutathione, γGC: γ-glutamyl cysteine, Glu: Glutamate. (b–e) A549 cells expressing scramble (SCR), NRF2 or PHGDH shRNAs were grown in the presence of U-13C-glucose for 24 hours and metabolites were extracted and analysed by LC/MS. (b) Analysis of 13C-labelling on the glycine component of glutathione. (c) Analysis of 13C-labeling on the purine metabolite adenosine monophosphate (AMP). (d,e) Analysis of glutathione (d) and AMP (e) labelling in serine high cell lines. Cells were labelled with 13C-glucose for 24 or 48 hours as indicated. (b,d) PHGDH-derived serine is incorporated into glutathione through the generation of glycine and cysteine. M+0 denotes no carbons labelled, M+2 denotes labelling on the glycine moiety, M+3 denotes labeling on the cysteine moiety, and M+5 denotes labeling on both glycine and cysteine. M+5 labeling was not observed. (c,e) M+5 labelling occurs following ribose-5-phosphate labelling via the pentose phosphate pathway. M+7 labelling is the result of ribose labelling plus either glycine or formyl-THF labelling in the purine ring. M+9 labelling is the result of ribose labelling plus either glycine and formyl-THF labelling in the purine ring. M+0 has no labelled carbons. (f–g) LC/MS analysis of total metabolite levels in the nucleotide (f) and transsulfuration (g) pathways. (h) NADPH/NADP+ ratios 4 days after PHGDH knockdown. Results are the average of 3 biological replicates.