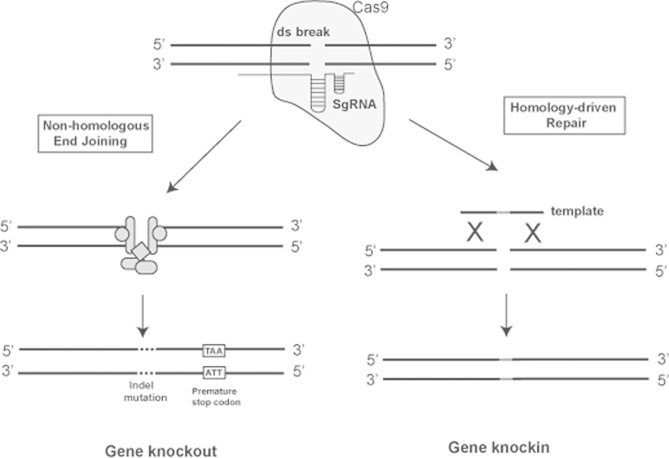
Fig. 4.
Gene editing using CRISPR/Cas. The CRIPSR/Cas system requires three components: a CRISPR-associated nuclease, for example Cas9; a single guide RNA (SgRNA) consisting of a guide sequence that binds the target DNA, a scaffold sequence for Cas9 binding and a terminating hairpin; and for gene knockin an oligo template containing the sequence to be inserted. The Cas9 is targeted to a specific genomic site by the SgRNA, where it induces a double strand break. In the absence of a repair template this break is repaired by non-homologous end joining, an error prone mechanism that leads to insertion/deletion (indel) mutations, which in turn cause frameshifts and the occurrence of premature stop codons, thereby leading to gene knockouts. If a DNA repair template is present the double strand break is repaired by a homology-driven repair mechanism, using this template, therefore genetic information can be knocked-in by including, for example, gain-of-function point mutations in the inserted template.