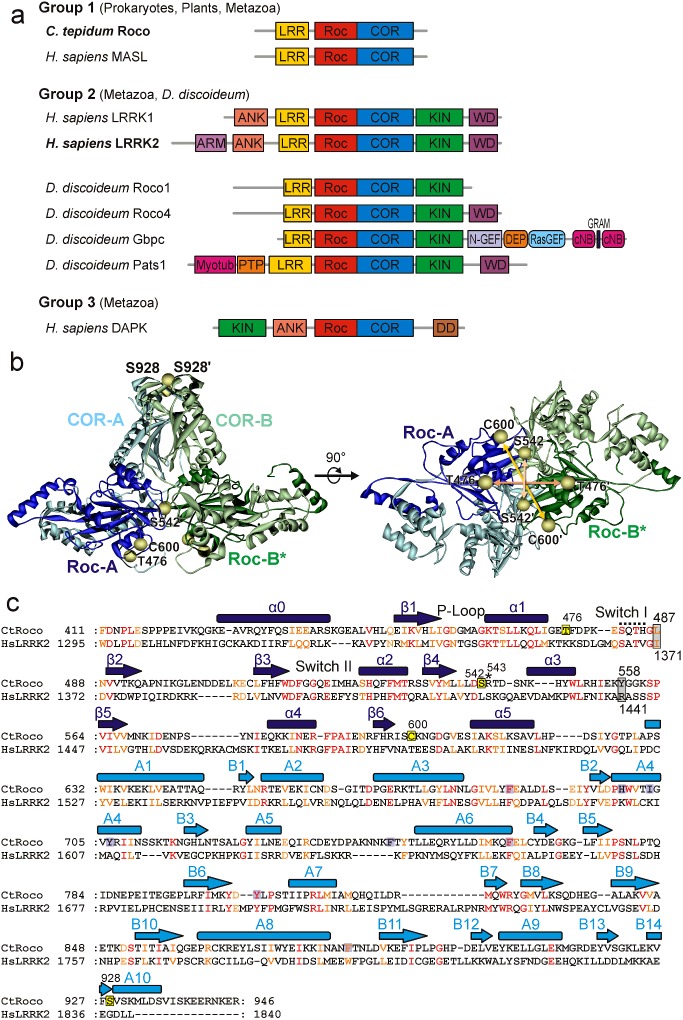
Figure 1. The Roco protein family.
(a) Domain topology of the Roco family proteins. The domains are ankyrin repeats (ANK), armadillo repeats (ARM), cyclic nt-binding domain (cNB), COR, death domain (DD), dishevelled, egl–10 and pleckstrin (DEP), Rab-like GTPase activators and myotubularins (GRAM), LRR, kinase (KIN), N-terminal motif of RasGEF (N-GEF), protein tyrosine phosphatase (PTP), RasGEF and Roc. (b) Model of the RocCOR dimer in two different orientations separated by 90°, with residues replaced by cysteine (except for Cys600) and subsequently labelled with MTSSL marked by spheres at the positions of their Cα atoms. The different protomers are shown in blue (light blue: COR-A, dark blue: Roc-A) and green (light green COR-B, dark green: Roc-B) respectively. The model has been created from the crystal structure of the C. tepidum RocCOR construct (pdb: 3DPU). The missing Roc-B domain in the X-ray structure was modelled into a position analogous to Roc-A. Loop regions not resolved in the structural model were also modelled (see ‘Materials and Methods’ for details). (c) Sequence alignment and secondary structure assignment of the RocCOR tandem for C. tepidum Roco and human LRRK2. Conserved residues are shown in red (identical amino acids) and orange (similar amino acids). Positions where the Parkinson mutations addressed in the present study appear in LRRK2 are marked by grey boxes. Spin-label positions are indicated by yellow boxes.